Table 1. Significant predictors of viral structural gene substitution rates.
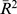
|
Predictor | β (95% CI) | Significance | |
| 1 | 0.73 | Neurons | −0.80 (−1.01, −0.59) | <0.0001 |
| Leukocytes | −0.56 (−0.80, −0.33) | <0.0001 | ||
| Hepatocytes | −0.24 (−0.40, −0.08) | 0.0004 | ||
| Endothelial cells | −0.18 (−0.31, −0.05) | 0.0007 | ||
| 2 | 0.73 | Epithelial cells | 1.13 (0.83, 1.43) | <0.0001 |
| Leukocytes | 0.53 (0.33, 0.73) | <0.0001 | ||
| 3 | 0.73 | Neurons | −0.39 (−0.53, −0.24) | <0.0001 |
| Epithelial cells | 0.58 (0.34, 0.82) | <0.0001 | ||
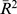
|
Predictor | β (95% CI) | Significance | |
| 1 | 0.73 | Neurons | −0.80 (−1.01, −0.59) | <0.0001 |
| Leukocytes | −0.56 (−0.80, −0.33) | <0.0001 | ||
| Hepatocytes | −0.24 (−0.40, −0.08) | 0.0004 | ||
| Endothelial cells | −0.18 (−0.31, −0.05) | 0.0007 | ||
| 2 | 0.73 | Epithelial cells | 1.13 (0.83, 1.43) | <0.0001 |
| Leukocytes | 0.53 (0.33, 0.73) | <0.0001 | ||
| 3 | 0.73 | Neurons | −0.39 (−0.53, −0.24) | <0.0001 |
| Epithelial cells | 0.58 (0.34, 0.82) | <0.0001 |
For each ANCOVA analysis, the overall adjusted R
2 (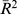 ) of the model is given along with significant predictor variables (P<0.01) and their standardized coefficients (β) with 95% confidence intervals (CIs). In the first ANCOVA, the base levels were epithelial target cells, fecal-oral/respiratory transmission route, acute/persistent infection, species-specific host range, and dsRNA genome architecture. In the second ANCOVA, the base levels were neural target cells, bites/scratches transmission route, persistent infection, order-specific host range, and (−)ssRNA genome architecture. In the third ANCOVA, the base levels were leukocyte target cells, respiratory/vertical transmission route, acute infection, family-specific host range, and (+)ssRNA genome architecture.
) of the model is given along with significant predictor variables (P<0.01) and their standardized coefficients (β) with 95% confidence intervals (CIs). In the first ANCOVA, the base levels were epithelial target cells, fecal-oral/respiratory transmission route, acute/persistent infection, species-specific host range, and dsRNA genome architecture. In the second ANCOVA, the base levels were neural target cells, bites/scratches transmission route, persistent infection, order-specific host range, and (−)ssRNA genome architecture. In the third ANCOVA, the base levels were leukocyte target cells, respiratory/vertical transmission route, acute infection, family-specific host range, and (+)ssRNA genome architecture.
