Background: The bone marrow environment secretes OPG and can thereby prevent TRAIL-induced apoptosis in multiple myeloma.
Results: rhTRAIL D269H/E195R shows a decreased affinity to OPG and induces apoptosis in multiple myeloma cells in the presence of OPG.
Conclusion: Apoptosis induction by rhTRAIL D269H/E195R is not inhibited by OPG.
Significance: rhTRAIL D269H/E195R can be used as potential therapy for multiple myeloma.
Keywords: Drug Design, Multiple Myeloma, Surface Plasmon Resonance (SPR), Trail, Tumor Microenvironment
Abstract
The bone marrow microenvironment provides important signals for the survival and proliferation of hematopoietic and malignant cells. In multiple myeloma, plasma cells are surrounded by stromal cells including osteoblasts. These stromal cells protect multiple myeloma cells from apoptosis induced by chemotherapeutic agents. Osteoprotegerin (OPG), a soluble receptor of the cytokine TNF-related apoptosis-inducing ligand (TRAIL), is secreted by osteoblasts and has been implicated in the prevention of cell death induced by TRAIL in malignant cells. Previously, we have designed death receptor-specific TRAIL variants that induce apoptosis exclusively via one of its death receptors. Here, we have studied in detail the interaction between recombinant human (rhTRAIL) variants and OPG. We show that a DR5-specific variant (rhTRAIL D269H/E195R) displays a significantly decreased affinity to OPG. Furthermore, this rhTRAIL variant shows a much higher activity when compared with rhTRAIL WT and retains its effectiveness in inducing cell death in multiple myeloma cell lines, in the presence of OPG secreted by stromal cells. We also demonstrate that stromal cells are largely insensitive to high concentrations of this rhTRAIL variant. In conclusion, rhTRAIL D269H/E195R is a potential therapy for multiple myeloma due to its high effectiveness and diminished binding to OPG.
Introduction
Multiple myeloma (MM)5 is a hematological malignancy characterized by clonal expansion of B cells within the bone marrow, which can result in osteolytic lesions, anemia, and immunosuppression. MM is in general an incurable disease when treated with conventional chemotherapy consisting of melphalan, prednisone, and proteasome inhibitors (1, 2). In many patients a relapse of the disease is observed, which suggests that a subpopulation of MM cancer stem cells is chemoresistant (2–4). These MM cancer stem cells have the ability to self-renew and expand (5, 6). The bone marrow microenvironment is essential for the survival and proliferation of myeloma stem cells, and it has been suggested that the microenvironment provides important cues in mediating drug resistance (4, 7, 8). Therefore, it is crucial to unravel the molecular mechanisms regulating microenvironment-mediated drug resistance to obtain novel therapeutic solutions.
Recently, it has been shown that tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL), a cytokine that selectively kills a variety of cancer cells, whereas leaving normal cells unharmed (9, 10), is able to efficiently induce apoptosis in multiple myeloma cell lines both in vitro and in vivo (11–13). Importantly, stem cell-enriched CD138− myeloma cells are sensitive to rhTRAIL treatment in combination with doxorubicin (14).
TRAIL activates the extrinsic pathway of apoptosis upon binding to its cognate surface death receptors 4 and 5 (DR4 and DR5). Ligand-induced receptor oligomerization of the receptors allows the assembly of the death-inducing signaling complex (DISC). Death-inducing signaling complex activation can then lead to the induction of apoptosis via the activation of a caspase cascade. However, the regulation of TRAIL-induced apoptosis is complex as TRAIL can also bind to the surface decoy receptors 1 and 2 (DcR1 and DcR2), both lacking an intact or a functional death domain, therefore preventing TRAIL-induced apoptosis. The soluble receptor osteoprotegerin (OPG) is also a binding partner of TRAIL. OPG is a soluble receptor that is secreted by osteoblasts residing in the bone marrow (15–17). This receptor has been shown to be involved in bone remodeling by binding to TNF superfamily-related protein receptor activator of NF-κB ligand (RANKL). This binding competes with RANKL binding to its surface receptor RANK, which is required for the maturation and activity of bone-absorbing osteoclasts (17). The promiscuous behavior of TRAIL and the presence of OPG within the bone marrow could therefore potentially compete with the association between TRAIL and its death-inducing receptors and interfere in TRAIL-mediated cell death of myeloma cells.
Previously, we developed death receptor-specific inducing rhTRAIL variants (18, 19). These variants trigger apoptosis specifically via either death receptor 4 (rhTRAIL 4C7 (G131R/R149I/N199R/K201H/S159R/S215D) or the death receptor 5 (rhTRAIL D269H/E195R). In this study, the impact of OPG in TRAIL-mediated apoptosis using rhTRAIL WT and death receptor-specific variants was further investigated in multiple myeloma cells, in the context of OPG released by their tumor microenvironment. We demonstrate that the lowered binding of a DR5-specific variant to OPG makes this variant insensitive to the interference in apoptosis mediated by this decoy receptor. This makes the rhTRAIL D269H/E195R variant a promising agent for therapeutic intervention in multiple myeloma tumors.
EXPERIMENTAL PROCEDURES
Determination of Receptor Binding by Surface Plasmon Resonance (SPR)
SPR buffers, regeneration solutions, and sensor chips were purchased from GE Healthcare. Protein A from Staphylococcus aureus was purchased from Sigma; receptor-Fc fusion OPG was from R&D Systems. Protein A was directly immobilized to all flow cells using a C1 sensor chip in a Biacore 3000 (in 10 mm NaAc, pH 4.5), and the primary amine coupling was performed according to the manufacturer's instructions (GE Healthcare). Experiments were carried out at 37 °C and a flow rate of 30 μl/min using HBS-P as running and dilution buffer (10 mm HEPES, pH 7.4, 150 mm NaCl, 0.005% (v/v) surfactant P20; GE Healthcare). After capturing ∼80 response units of OPG-Fc receptor at a high flow rate, a method comprising a single-cycle approach, where the analyte is injected with increasing concentrations of rhTRAIL WT and variants on a single cycle, was performed. A total volume of 50 μl of rhTRAIL was injected per concentration, and correction of all binding curves was performed by so-called double referencing, i.e. subtraction of the data of the “empty” flow cell 1 followed by subtraction of the data from a run buffer injection cycle.
ELISA and Competitive ELISA Assays
Nunc MaxiSorp plates were coated for 1 h with OPG-Fc (100 ng/well) in 0.1 m sodium carbonate/bicarbonate buffer (pH 8.6), and the remaining binding places were subsequently blocked with 2% BSA for 1 h. After washing for six times with Tris-buffered saline/0.5% Tween 20 (TBST) (pH 7.5), serial dilutions of rhTRAIL WT and rhTRAIL D269H/E195R (0–2000 ng/well) were added and incubated at 37 °C for 1 h. After washing with TBST, a 1:200 dilution of goat anti-TRAIL antibody (R&D Systems) was added and incubated for 1 h at room temperature, and after washing six times with TBST, subsequently incubated with a 1:1000 dilution of a horseradish peroxidase-conjugated swine anti-goat antibody. After washing six times with TBST, 100 μl of 3,3′,5,5′-tetramethylbenzidine solution (Calbiochem) was added, and after 5 min, the reaction was quenched with 100 μl of 1 m sulfuric acid. The absorbance was measured at 450 nm on a microplate reader (Thermo Labsystems). For the competitive enzyme-linked immunosorbent assay (ELISA) assay, wells were coated with DR5-Fc as described above. Serial dilutions of soluble OPG-Fc (0–1000 ng/well) and rhTRAIL WT or rhTRAIL D269H/E195R (10 ng/well) in PBS (pH 7.4) were preincubated for 1 h at 37 °C, added to the wells, and further incubated for 1 h at 37 °C. Binding of rhTRAIL WT or rhTRAIL D269H/E195R to immobilized DR5-Fc with 0 ng/well of OPG-Fc was taken as 100%, and binding at other concentrations of OPG-Fc was calculated relative to 0 ng/well of OPG-Fc.
Cell Lines and Reagents
The human multiple myeloma cell lines RPMI 8226 and U266 were cultured in RPMI with 10% FCS and penicillin/streptomycin. The osteosarcoma osteoblast-like cell line MG63 was cultured in DMEM-F12 containing 10% FCS and penicillin/streptomycin. The osteosarcoma osteoblast-like cell line SaOS2 was cultured in DMEM containing 10% FCS and penicillin/streptomycin. rhTRAIL WT, rhTRAIL 4C7, and rhTRAIL D269H/E195R variants were expressed and purified as described previously (18, 19). Recombinant human OPG and monoclonal anti-human OPG antibodies were purchased from R&D Systems (185-OS and MAB805).
Cell Line Treatment
RPMI 8226 and U266 cells were seeded, and after 24 h, cells were incubated with various concentrations of rhTRAIL WT or variants with or without rhOPG. RhTRAIL WT, rhTRAIL 4C7, or rhTRAIL D269H/E195R was preincubated with rhOPG for 15 min at 37 °C. TRAIL combined to OPG was then added to RPMI 8226 or U266 and cells and further incubated for 24 h. Cell death was assessed by the CellTiter 96® AQueous non-radioactive cell proliferation assay (MTS) (Promega), by annexin V staining, and by acridine orange staining.
MTS Assay
Briefly, after incubating the cells for 24 h at 37 °C using 96-well plates, 20 μl of the MTS reagent was added to 100 μl of treated cells, and cell viability was determined by measuring the absorption at 490 nm using a microplate reader (Thermo Labsystems), according to the manufacturer's instructions. For the co-culture assay, MG63 cells were seeded, and after 48 h, U266 cells were resuspended in conditioned medium and added to MG63 cells. Thereafter, cells were incubated with various concentrations of rhTRAIL WT or rhTRAIL D269H/E195R with or without rhOPG.
Annexin V Staining
After 24 h of incubation at 37 °C in a 24-well plate, U266 cells were washed in calcium buffer (10 mm HEPES/NaOH, pH 7.4, 140 mm NaCl, 2.5 mm CaCl2), and 3 μl (in a total volume of 60 μl of PBS) of annexin-V-FITC antibody (IQ-products IQ-120F) was added for 20 min at 4 °C. Cells were washed with calcium buffer and analyzed by FACSCalibur.
Acridine Orange
After 24 h of incubation, acridine orange was added to the cells for 15 min. Cells with condensed DNA and modified morphology were counted as apoptotic cells.
Receptor Expression Analysis
U266 and RPMI 8226 cells were grown, and after 24 h, cells were incubated with mouse IgG1 isotype control (Dako, X0931), α-TRAIL-R1 (Enzo Life Sciences, ALX-804-297A), α-TRAIL-R2 (Enzo Life Sciences, ALX-804-298A), α-TRAIL-R3 (Enzo Life Sciences, ALX-804-344A), or α-TRAIL-R4 (Enzo Life Sciences, ALX-804-299A). After washing the cells, cells were incubated with donkey anti-mouse FITC (Jackson ImmunoResearch Laboratories, 715-095-150) at 4 °C for 1 h. Cells were washed, and fluorescence-activated cell sorting (FACS) analysis was performed on a FACSCalibur (BD Biosciences). Data were further analyzed using FlowJo 7.6.1.
Determination of OPG Secretion by MG63 and SaOS2 Cell Lines
To determine the amount of OPG secreted by the osteosarcoma osteoblast-like MG63 and SaOS2 cells, both cell lines were seeded at 80% confluency, and 48 h later, conditioned medium from these cells was harvested. Total OPG concentration was measured using the human OPG ELISA kit (R&D Systems, DY805) following the manufacturer's instructions. In short, a 96-well plate was coated overnight at room temperature with mouse anti-human OPG capturing antibody. On the next day, the 96-well plate containing OPG antibody was washed and blocked with BSA, and conditioned medium was added. After 2 h of incubation, wells were washed and incubated with a biotinylated goat anti-human OPG detection antibody. Subsequently, OPG concentration was measured by the addition of streptavidin-HRP conjugate and the HRP-substrate solution. As a standard, recombinant human OPG was added at various concentrations to quantify the OPG concentration secreted by the cells.
Western Blotting
For antigen detection, membranes were incubated with antibodies to actin (1:1000; Santa Cruz Biotechnology) and cleaved caspase 8 (1:1000; Cell Signaling Technologies) overnight at 4 °C followed by a 2-h incubation at room temperature with appropriate secondary antibodies (1:10000; Invitrogen).
RESULTS
RhTRAIL D269H/E195R Shows a Lowered Binding to OPG-Fc When Compared with rhTRAIL WT and 4C7
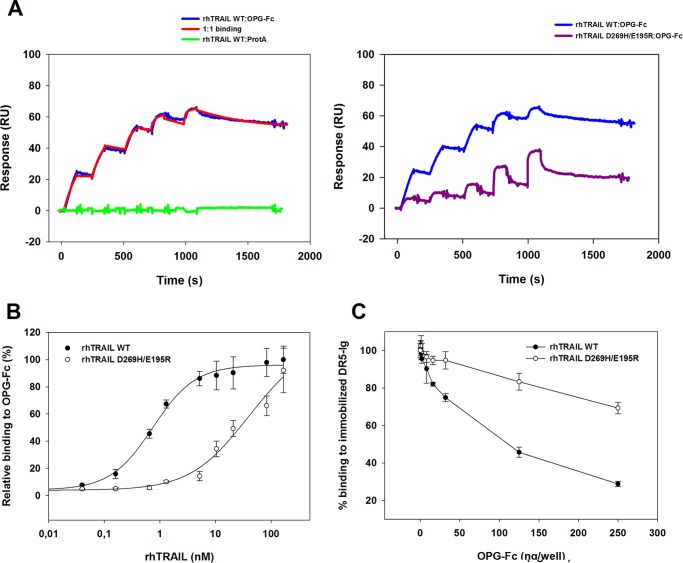
Analysis of the affinity between rhTRAIL WT or the death receptor-specific variants and OPG was performed using SPR and ELISA assays. Recently, variations in receptor density and sensor chip type have allowed us to manipulate the stoichiometry of the formed receptor-ligand complexes by using SPR and to determine the rate constants by describing the binding of trimeric rhTRAIL to one single receptor molecule (20). We have performed kinetic characterization of the interaction between rhTRAIL WT/death receptor-specific variants and OPG-Fc using a single-cycle approach to determine the rate constants describing the binding of trimeric TRAIL to one OPG receptor molecule. Using this method, the analyte was injected with increasing concentrations on a single cycle, and the surface was not regenerated between sequential injections. Single-cycle kinetics enabled us to reduce the time spent on each assay, whereas still allowing kinetic analysis of the molecular interaction between TRAIL and OPG. The capture method in which the OPG-Fc molecules are bound by immobilized protein A allowed us to calculate that at the low density of 80 response units, solely a trimer-monomer complex was formed (expected response was 80 × 58.5/71 = 65.9 response units; 58.5 kDa corresponds to the molecular mass of TRAIL, and 71 kDa is the predicted molecular mass for one arm of the disulfide linked homodimeric OPG chimera). The interaction data obtained for rhTRAIL WT:OPG-Fc could be accurately fitted using a 1:1 binding, resulting in a dissociation constant (KD) of 0.9 nm (Fig. 1A), with ka = 1.0 × 106 m−1 s−1 and kd = 1.02 × 10−3 s−1. Interestingly, rhTRAIL 4C7 was able to bind OPG-Fc with similar affinity as observed for rhTRAIL WT, resulting in a KD of 1.4 nm (supplemental Fig. 1). Analysis of rhTRAIL D269H/E195R showed a considerable decline in binding to OPG-Fc when compared with rhTRAIL WT (Fig. 1A). Although no satisfactory fit could be obtained with the available software, clearly both the association and the dissociation rate constants are affected. In addition, pre-steady state analysis, performed as described in Refs. 18, 21, and 22 by reading the response values after the end of the injection and plotting these as a function of the rhTRAIL concentrations, indicate a nearly 70-fold decrease in affinity for rhTRAIL D269H/E195R:OPG-Fc when compared with rhTRAIL WT:OPG (18). ELISA assays further confirmed the decrease in affinity between rhTRAIL D269H/E195R and OPG-Fc with regard to the affinity measured for rhTRAIL WT and rhTRAIL 4C7 to OPG-Fc (Fig. 1B, supplemental. Fig. 1). Analysis of the interaction between rhTRAIL WT and OPG-Fc is consistent with the values previously obtained by SPR for rhTRAIL WT binding to OPG-Fc (KD = 0.75 nm) (Table 1). Furthermore, the variant D269H/E195R showed an ∼89-fold decrease in apparent affinity (KD = 66.8 nm) to OPG-Fc when compared with rhTRAIL WT as assessed by ELISA. We further evaluated the binding preference of the mutant rhTRAIL D269H/E195R by performing a competitive ELISA assay using coated DR5-Fc and competitive soluble OPG-Fc (Fig. 1C). Soluble OPG-Fc was very efficient in reducing the binding of rhTRAIL WT to immobilized DR5-Fc, in contrast to the results obtained for rhTRAIL D269H/E195R, which was much less sensitive to competition by OPG-Fc. Notably, the mutant still showed ∼70% binding efficiency to DR5-Fc even when competing with 2500 ng/ml OPG-Fc (Fig. 1C). Taken together, these results indicate a decreased affinity of rhTRAIL D269H/E195R toward OPG, when compared with the affinity constants determined for rhTRAIL WT, and a lowered capacity of OPG-Fc to compete for DR5-Fc binding by this mutant, when compared with both rhTRAIL WT and rhTRAIL 4C7.
FIGURE 1.
Receptor binding of rhTRAIL WT and rhTRAIL D269H/E195R as determined by SPR and ELISA. A, SPR sensorgrams for trimer-monomer complex formation of rhTRAIL WT or rhTRAIL D269H/E195R (6.25–100 nm; left to right) and OPG-Fc. Using single-site kinetics, binding of rhTRAIL WT (A, left and right) and rhTRAIL D269H/E195R (A, right) to ∼70 response units (RU) of OPG-Fc captured by protein A was assessed using a C1 sensor chip and a flow rate of 30 μl/min in HBS-P. A flow channel containing only immobilized protein A subtracted to a run buffer injection cycle was used as control (A; green). The data were fitted using a titration kinetics 1:1 binding with drift in BIAevaluation 4.1. B, receptor binding of rhTRAIL WT and rhTRAIL D269H/E195R to OPG-Fc as determined by ELISA. Receptor binding was calculated relative to the response of rhTRAIL WT at 110 nm. C, competitive ELISA by TRAIL receptors of rhTRAIL WT and rhTRAIL D269H/E195R binding to immobilized DR5-Ig receptor using soluble OPG-Fc as a competitor. Ten nanograms/well of rhTRAIL WT or rhTRAIL D269H/E195R was preincubated with 0–250 ng/well of OPG-Fc for 1 h. Preincubated solutions were added to microtiter plates coated with DR5-Ig. Binding of the variants at various concentrations of soluble receptor toward the immobilized DR5-Fc was calculated relative to the value measured on the presence of 0 ng/well of soluble receptor. The data are the mean ± S.D. of three independent experiments.
TABLE 1.
Apparent OPG-Fc binding affinities of rhTRAIL WT and rhTRAIL D269H/E195R as determined by ELISA
Apparent KD values for OPG-Fc were calculated using a four-parameter fitting tool.
| Protein | KD |
|---|---|
| nm | |
| rhTRAIL WT | 0.75 ± 0.09 |
| rhTRAIL 4C7 | 0.67 ± 0.08 |
| rhTRAIL D269H/E195R | 66.8 ± 6.02 |
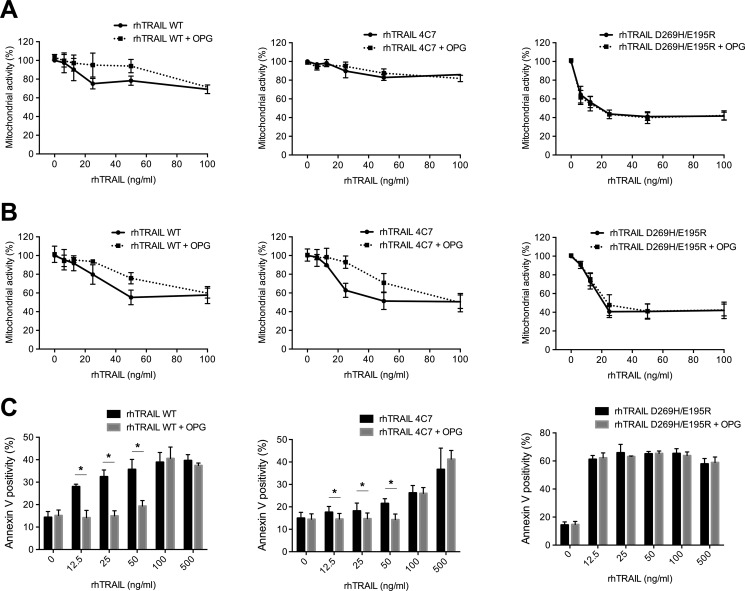
Activity of the rhTRAIL-D269H/E195R Variant Is Unaffected in the Presence of OPG and Efficiently Induces Cell Death in Multiple Myeloma Cell Lines
The biological activity of rhTRAIL WT and the two variants rhTRAIL 4C7 and D269H/E195R was measured in the multiple myeloma cell lines RPMI 8226 and U266. The DR5-specific variant was the most potent inducer of apoptosis in these cell lines, when compared with rhTRAIL 4C7 and rhTRAIL WT, as measured by MTS and annexin V staining (Fig. 2, A–C). To assess the biological consequences of the altered binding kinetics of DR4- and DR5-specific variants toward OPG, we analyzed the survival of the multiple myeloma cell lines RPMI 8226 and U266 upon the addition of rhTRAIL WT, rhTRAIL 4C7, and rhTRAIL D269H/E195R in the presence of 100 ng/ml rhOPG. Consistent with the decreased binding affinity of the rhTRAIL D269H/E195R variant to OPG, the addition of rhOPG did not shield these two cell lines from apoptosis induced by this variant, whereas the same cells became less sensitive to apoptosis when treated with rhTRAIL WT (Fig. 2, A–C). Upon the addition of rhOPG, RPMI 8226 cells treated with rhTRAIL 4C7 became less sensitive to apoptosis mediated by this variant, suggesting that OPG can also interfere with rhTRAIL 4C7 apoptosis induction. Interestingly, statistically significant differences in apoptosis were also observed in U266 cells upon the addition of low concentrations of rhTRAIL 4C7 in the presence of rhOPG. The less pronounced decrease in activity is most likely due to the very low activity of TRAIL 4C7 at these concentrations, consistent with the low DR4 expression of this cell line (supplemental Fig. 2). Furthermore, we confirmed that rhTRAIL WT and the variants activate the initiator caspase-8 (supplemental Fig. 3). The addition of OPG partially rescued U266 cells from apoptosis induced by rhTRAIL WT and rhTRAIL 4C7, consistent with a reduced activation of caspase-8 in cells treated with these proteins. In contrast, no differences in caspase-8 activation could be observed upon the addition of OPG to rhTRAIL D269H/E195R-treated cells, confirming that OPG does not reduce rhTRAIL D269H/E195R-induced caspase-8 activation.
FIGURE 2.
Sensitivity of multiple myeloma cells lines to rhTRAIL D269H/E195R variant is unchanged upon the addition of OPG. A and B, U266 cells (A) and RPMI 8226 cells (B) were treated with rhTRAIL WT, rhTRAIL 4C7, or the rhTRAIL D269H/E195R for 24 h in the presence or absence of 100 ng/ml OPG. On the next day, cell viability (mitochondrial activity) was evaluated using an MTS assay. C, apoptosis induced by rhTRAIL WT, rhTRAIL 4C7, and rhTRAIL D269H/E195R in U266 cells was quantified by annexin V staining. The data are the mean ± S.D. of three independent experiments.
Death Receptor Expression Influences the Sensitivity to TRAIL-mediated Apoptosis
We questioned whether the difference in sensitivity of these multiple myeloma cell lines could be related to the varied expression of the death receptors on the surface of the cells. The activities shown for the DR4-specific rhTRAIL 4C7 and DR5-specific rhTRAIL D269H/E195R variants correlate with the surface expression of death receptors DR4 and DR5 on the surface of these myeloma cell lines. RPMI 8226 cells were sensitive to both DR4-mediated and DR5-mediated apoptosis and expressed both DR4 as well as DR5, whereas U266 cells (sensitive to the rhTRAIL D269H/E195R variant and only mildly sensitive to rhTRAIL 4C7) expressed significantly higher levels of DR5 when compared with DR4 (supplemental Fig. 2, supplemental Table 1).
OPG Secreted by Osteoblast-like Cells Protects U266 cells from Apoptosis Mediated by rhTRAIL WT, but Not by rhTRAIL D269H/E195R
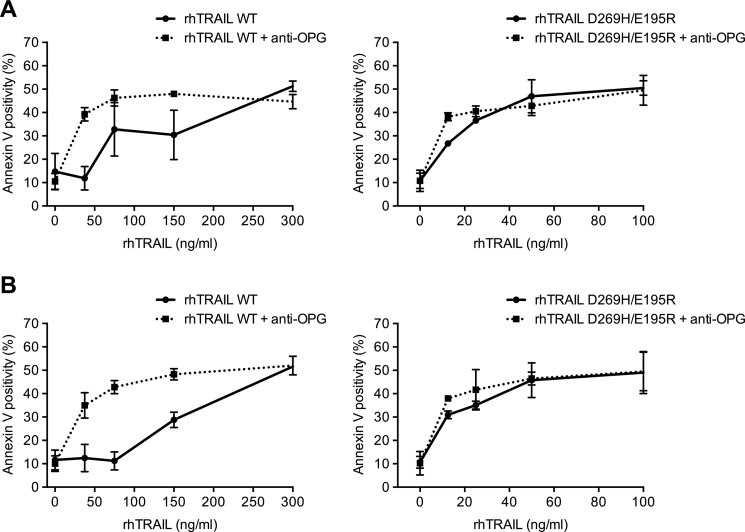
OPG is secreted by osteoblasts and by marrow stromal cells. Medium conditioned by the human osteosarcoma osteoblast cell lines MG63 or SaOS2 for 2 days contained, respectively, 260 and 7 ng/ml OPG as measured by ELISA (Table 2). In contrast to these osteosarcoma cell lines, the multiple myeloma cells RPMI 8226 and U266 did not secrete OPG (below detection level of 0.2 ng/ml). To investigate whether rhTRAIL D269H/E195R retained its efficacy in the presence of OPG secreted by these osteoblast-like cells, we set up a co-culture system of the myeloma U266 cells and the osteoblast-like MG63 cells. To verify that TRAIL-induced apoptosis does not occur in MG63 cells and therefore interferes with apoptosis measurement in the multiple myeloma cell lines tested, we performed acridine orange staining using different concentrations of both rhTRAIL WT and variant (Fig. 3). Our results indicate that MG63 cells are not sensitive to either rhTRAIL WT or rhTRAIL D269H/E195R, even upon the addition of a very high concentration of rhTRAIL WT or rhTRAIL D269H/E195R (1000 ng/ml). Upon the addition of conditioned medium from MG63 cells to the U266 myeloma cells, U266 cells showed a clear reduction in cell death induced by rhTRAIL WT (Fig. 4A). However, conditioned medium did not affect apoptosis levels induced by rhTRAIL D269H/E195R. To test whether the effect of the conditioned medium was mediated by OPG, 2 μg/ml of a monoclonal anti-human OPG antibody was added to this culture. The addition of anti-OPG reversed the effects of the conditioned medium observed in the U266 cells upon treatment with rhTRAIL WT, demonstrating that OPG was responsible for this reduction in apoptosis (Fig. 4A). Furthermore, a similar reduction in apoptosis was observed when U266 cells were co-cultured with osteosarcoma MG63 cells. Equal apoptosis induction activities were observed for rhTRAIL D269H/E195R when U266 cells were co-cultured with MG63 cells in the presence or absence of anti-OPG, whereas U266 cells became resistant to rhTRAIL WT-mediated apoptosis due to OPG secreted from the MG63 cells (Fig. 4B).
TABLE 2.
Levels of OPG secretion into the medium of MG63, SaOS2, U266, and RPMI 8226 cells
| Cell line | OPG secretion |
|---|---|
| MG63 | 260 ng/ml |
| SaOS2 | 7 ng/ml |
| U266 | <0.2 ng/ml (detection limit) |
| RPMI 8226 | <0.2 ng/ml (detection limit) |
FIGURE 3.
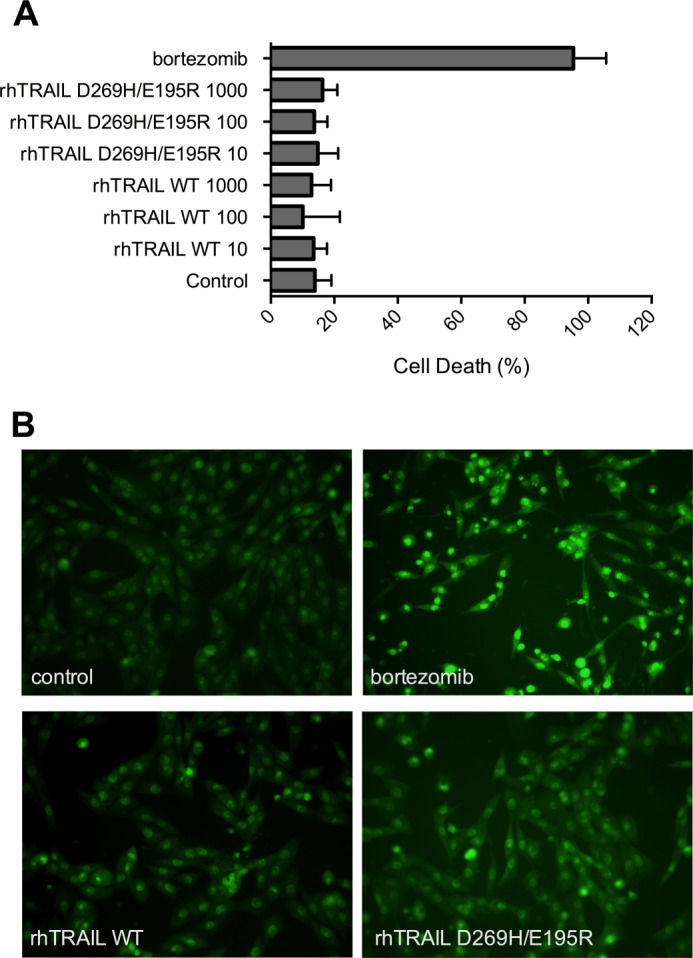
MG63 cells are resistant to rhTRAIL WT and rhTRAIL D269H/E195R. A, quantification of apoptotic MG63 cells counted by acridine orange staining, upon treatment with 50 μm bortezomib and 10, 100, and 1000 ng/ml rhTRAIL WT or rhTRAIL D269H/E195R. The data are the mean ± S.D. of three independent experiments. B, illustration of acridine orange-stained cells in control MG63 cells, in MG63 cells treated with 50 μm bortezomib (positive control), and in MG63 cells treated with 1 μg/ml rhTRAIL WT or rhTRAIL D269H/E195R.
FIGURE 4.
Cell death induced by rhTRAIL D269H/E195R is not inhibited by OPG secreted by stromal MG63 cells in contrast to rhTRAIL WT. A, conditioned medium (2 days) was added to U266 cells, and cells were incubated with rhTRAIL WT or rhTRAIL D269H/E195R for 24 h. OPG present in the medium was neutralized by the addition of an OPG-neutralizing antibody prior to treatment. B, U266 cells in co-culture with MG63 cells were incubated with rhTRAIL WT (0–300 ng/ml) or rhTRAIL D269H/E195R (0–100 ng/ml), and after 24 h, apoptosis of suspension cells was measured by annexin V staining. The data are the mean ± S.D. of two independent experiments.
DISCUSSION
Multiple myeloma is a plasma B-cell disorder that still has an unfavorable prognosis using conventional chemotherapies. Successful treatment of multiple myeloma and other hematological malignancies does not only depend on the efficacy of the therapeutic agents but is further dictated by the bone microenvironment surrounding the multiple myeloma cells. This bone microenvironment consists of several cells supporting the proliferation and survival of normal and malignant hematopoietic (stem) cells (3, 4). Furthermore, the presence of these supporting cells and their secreted cytokines might protect against various chemotherapies and contribute to chemoresistance (5, 6). OPG is secreted by bone marrow cells and prevents excessive bone resorption by osteoclasts. Maturation of osteoclasts is inhibited by OPG via binding to RANKL, the ligand that activates osteoclasts via the RANK receptor. However, OPG might also function as a paracrine survival factor of myeloma cells by binding to TRAIL, and thereby it could prevent induction of apoptosis in these malignant cells. TRAIL is a promising therapeutic agent that selectively induces apoptosis in tumor cells when compared with normal cells. It is of potential clinical relevance in multiple myeloma due to its ability to specifically target the MM cancer stem cells that are required for the development and relapse of the disease (14). To further improve the efficacy of rhTRAIL WT, we were recently successful in developing rhTRAIL variants that specifically target cancer cells via either the DR5 or the DR4 receptor and induce apoptosis in various tumor cells, with improved effectiveness when compared with rhTRAIL WT (18, 19). We have characterized the affinity of our most potent death receptor-specific inducing variants to OPG. Receptor binding experiments using SPR and ELISA assays demonstrate that rhTRAIL D269H/E195R displays a significantly lowered binding affinity to OPG-Fc when compared with rhTRAIL WT and 4C7 (Fig. 1, A and B, supplemental Fig. 1, Table 1). In addition, competitive ELISA assays reveal that soluble OPG-Fc is largely ineffective in competing for rhTRAIL D269H/E195R binding to DR5-Fc when compared with rhTRAIL WT (Fig. 1C). As different binding affinities of TRAIL to its receptors can influence TRAIL activity, the results presented here indicate that OPG binds with high affinity to TRAIL and competes in vitro for death receptor binding. This is in accordance to previous results obtained by Vitovski et al. (23), and in contrast with the results obtained by Truneh et al. (24), where OPG was able to compete with TRAIL binding to immobilized DR5 only at 4 °C but not at 37 °C. Although the apparent affinities obtained here are in agreement with or similar to the apparent dissociation constants reported for rhTRAIL WT binding to OPG by Gasparian et al. (25) and Emery et al. (26), when using SPR, they differ somewhat from those obtained by others (24). These differences in affinity may be related to the fact that apparent KD values can be substantially higher when equilibrium at low concentrations is not reached (20). Additionally, differences in the methodology used, including immobilization versus capturing techniques, or/and the preparation of the recombinant molecules used in these studies (e.g. introduction of tags that that can influence the oligomerization of ligands such as TRAIL) could influence the final affinity constants obtained. Importantly, tumor cells and osteoclasts producing OPG can prevent TRAIL-induced apoptosis of tumor cells (16, 17), indicating that OPG plays a regulatory role in TRAIL-induced apoptosis.
The residue Asp-269 plays an important role in conferring DR5 selectivity (18); however, it is not in direct contact with the DR5 receptor. Strikingly, this is not the case for the models of TRAIL in complex with DR4, DcR1, and DcR2, where Asp-269 of TRAIL directly interacts with Lys-120 of these receptors. In DR5, this residue is replaced by an aspartate. A sequence alignment performed on all the receptors of TRAIL reveals that OPG also contains a Lys at this position; consequently, it is plausible that the interference mediated by this mutation on OPG binding partially results from breaking this Asp-Lys interaction, as described previously for the receptors DR4, DcR1, and DcR2, and ultimately leads to a decreased affinity to the receptor OPG (18). To fully resolve the underlying mechanism of OPG binding to rhTRAIL WT and rhTRAIL D269H/E195R, the elucidation of the crystal structure of the complex between OPG and TRAIL, or a model of rhTRAIL in complex with OPG, making use of the recently identified structure of OPG in complex with RANKL (27, 28), might give us further insight into the binding characteristics of this complex. This could potentially function as a lead to further improve or design rhTRAIL variants that do not bind to OPG. The affinity results obtained and the likely role of OPG-mediated inhibition on TRAIL-mediated apoptosis (23) allowed us to propose that the variant rhTRAIL D269H/E195R could eventually bypass OPG-mediated TRAIL resistance in the bone marrow environment. To demonstrate that the differences in affinity are reflected in vitro, we demonstrate that rhTRAIL D269H/E195R retains its effectiveness in the presence of OPG, whereas OPG protects multiple myeloma cells from apoptosis induced by rhTRAIL WT and rhTRAIL 4C7 (Fig. 2). Analysis of the surface expression of death receptors on these cells correlates with the specific activities observed in these cell lines by DR-specific variants (supplemental Fig. 3). Interestingly, despite the high expression of DcR2, rhTRAIL D269H/E195R still induced high levels of apoptosis in the RPMI 8226 cells.
Using a co-culture experiment of myeloma U266 cells and osteoblast-like MG63 cells, we show that rhTRAIL D269H/E195R is insensitive to OPG secreted by MG63 cells, whereas the activity of rhTRAIL WT is impaired by the addition of OPG and rescued by the subsequent addition of anti-OPG (Fig. 4). Notably, the stromal MG63 cells were insensitive to high concentrations of both rhTRAIL WT and rhTRAIL D269H/E195R, and therefore these cells did not affect the quantification of apoptosis in our assays (Fig. 3).
To further evaluate the effect of decreased binding of rhTRAIL D269H/E195R to OPG and thereby increased efficacy when compared with rhTRAIL WT, in vivo validation will be required. Recently, it has been suggested that OPG does not prevent rhTRAIL WT-induced cell death in a mouse model (29). However, in that study, target cells themselves did overexpress OPG and were injected intratibially, which differs from the bone marrow microenvironment consisting of multiple myeloma cells and stromal cells. Furthermore, for therapeutic applications, low levels of rhTRAIL and the differential affinities observed for all its receptors may allow locally high concentrations of OPG (30) to take a more predominant role in preventing TRAIL-induced apoptosis in the context of the bone marrow environment. As such, OPG may play a more dominant role in protecting these cells from undergoing apoptosis in vivo than previously anticipated.
In conclusion, we have shown that the rhTRAIL D269H/E195R variant has a decreased binding affinity to OPG and that this variant is superior to rhTRAIL WT in targeting several multiple myeloma cells. This superior effectiveness is mediated by its primary effect on multiple myeloma cells, and it remains highly effective due to its decreased binding to OPG secreted by bone marrow. Thus, these results demonstrate that rhTRAIL D269H/E195R can effectively target multiple myeloma cells by bypassing a therapeutic interference mediated by their microenvironment, and improve the available options for therapeutic intervention in multiple myeloma tumors.

This article contains supplemental Figs. 1–3 and Table 1.
- MM
- multiple myeloma
- TRAIL
- TNF-related apoptosis-inducing ligand
- rhTRAIL
- recombinant human TRAIL
- OPG
- osteoprotegerin
- rhOPG
- recombinant human OPG
- DR
- death receptor
- DcR
- decoy receptor
- RANKL
- receptor activator of NF-κB ligand
- MTS
- 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt.
REFERENCES
- 1. Kyle R. A., Rajkumar S. V. (2004) Multiple myeloma. N. Engl. J. Med. 351, 1860–1873 [DOI] [PubMed] [Google Scholar]
- 2. Palumbo A., Anderson K. (2011) Multiple myeloma. N. Engl. J. Med. 364, 1046–1060 [DOI] [PubMed] [Google Scholar]
- 3. Jones R. J., Armstrong S. A. (2008) Cancer stem cells in hematopoietic malignancies. Biol. Blood Marrow Transplant. 14, Suppl. 1, 12–16 [DOI] [PubMed] [Google Scholar]
- 4. Valent P., Bonnet D., De Maria R., Lapidot T., Copland M., Melo J. V., Chomienne C., Ishikawa F., Schuringa J. J., Stassi G., Huntly B., Herrmann H., Soulier J., Roesch A., Schuurhuis G. J., Wöhrer S., Arock M., Zuber J., Cerny-Reiterer S., Johnsen H. E., Andreeff M., Eaves C. (2012) Cancer stem cell definitions and terminology: the devil is in the details. Nat. Rev. Cancer 12, 767–775 [DOI] [PubMed] [Google Scholar]
- 5. Huff C. A., Matsui W. (2008) Multiple myeloma cancer stem cells. J. Clin. Oncol. 26, 2895–2900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Matsui W. (2011) Perspective: A model disease. Nature 480, S58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Donnenberg V. S., Donnenberg A. D. (2005) Multiple drug resistance in cancer revisited: the cancer stem cell hypothesis. J. Clin. Pharmacol. 45, 872–877 [DOI] [PubMed] [Google Scholar]
- 8. Silvestris F., Cafforio P., Tucci M., Grinello D., Dammacco F. (2003) Upregulation of osteoblast apoptosis by malignant plasma cells: a role in myeloma bone disease. Br. J. Haematol. 122, 39–52 [DOI] [PubMed] [Google Scholar]
- 9. Gonzalvez F., Ashkenazi A. (2010) New insights into apoptosis signaling by Apo2L/TRAIL. Oncogene 29, 4752–4765 [DOI] [PubMed] [Google Scholar]
- 10. Yang A., Wilson N. S., Ashkenazi A. (2010) Proapoptotic DR4 and DR5 signaling in cancer cells: toward clinical translation. Curr. Opin. Cell Biol. 22, 837–844 [DOI] [PubMed] [Google Scholar]
- 11. Gazitt Y. (1999) TRAIL is a potent inducer of apoptosis in myeloma cells derived from multiple myeloma patients and is not cytotoxic to hematopoietic stem cells. Leukemia 13, 1817–1824 [DOI] [PubMed] [Google Scholar]
- 12. Lincz L. F., Yeh T. X., Spencer A. (2001) TRAIL-induced eradication of primary tumour cells from multiple myeloma patient bone marrows is not related to TRAIL receptor expression or prior chemotherapy. Leukemia 15, 1650–1657 [DOI] [PubMed] [Google Scholar]
- 13. Mitsiades C. S., Treon S. P., Mitsiades N., Shima Y., Richardson P., Schlossman R., Hideshima T., Anderson K. C. (2001) TRAIL/Apo2L ligand selectively induces apoptosis and overcomes drug resistance in multiple myeloma: therapeutic applications. Blood 98, 795–804 [DOI] [PubMed] [Google Scholar]
- 14. Vitovski S., Chantry A. D., Lawson M. A., Croucher P. I. (2012) Targeting tumour-initiating cells with TRAIL based combination therapy ensures complete and lasting eradication of multiple myeloma tumours in vivo. PLoS One 7, e35830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Locklin R. M., Croucher P. I., Russell R. G., Edwards C. M. (2007) Agonists of TRAIL death receptors induce myeloma cell apoptosis that is not prevented by cells of the bone marrow microenvironment. Leukemia 21, 805–812 [DOI] [PubMed] [Google Scholar]
- 16. Shipman C. M., Croucher P. I. (2003) Osteoprotegerin is a soluble decoy receptor for tumor necrosis factor-related apoptosis-inducing ligand/Apo2 ligand and can function as a paracrine survival factor for human myeloma cells. Cancer Res. 63, 912–916 [PubMed] [Google Scholar]
- 17. Holen I., Shipman C. M. (2006) Role of osteoprotegerin (OPG) in cancer. Clin. Sci. 110, 279–291 [DOI] [PubMed] [Google Scholar]
- 18. van der Sloot A. M., Tur V., Szegezdi E., Mullally M. M., Cool R. H., Samali A., Serrano L., Quax W. J. (2006) Designed tumor necrosis factor-related apoptosis-inducing ligand variants initiating apoptosis exclusively via the DR5 receptor. Proc. Natl. Acad. Sci. U.S.A. 103, 8634–8639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Reis C. R., van der Sloot A. M., Natoni A., Szegezdi E., Setroikromo R., Meijer M., Sjollema K., Stricher F., Cool R. H., Samali A., Serrano L., Quax W. J. (2010) Rapid and efficient cancer cell killing mediated by high-affinity death receptor homotrimerizing TRAIL variants. Cell Death Dis. 1, e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Reis C. R., van Assen A. H., Quax W. J., Cool R. H. (2011) Unraveling the binding mechanism of trivalent tumor necrosis factor ligands and their receptors. Mol. Cell. Proteomics 10, M110.002808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Reis C. R., van der Sloot A. M., Szegezdi E., Natoni A., Tur V., Cool R. H., Samali A., Serrano L., Quax W. J. (2009) Enhancement of antitumor properties of rhTRAIL by affinity increase toward its death receptors. Biochemistry 48, 2180–2191 [DOI] [PubMed] [Google Scholar]
- 22. Tur V., van der Sloot A. M., Reis C. R., Szegezdi E., Cool R. H., Samali A., Serrano L., Quax W. J. (2008) DR4-selective tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) variants obtained by structure-based design. J. Biol. Chem. 283, 20560–20568 [DOI] [PubMed] [Google Scholar]
- 23. Vitovski S., Phillips J. S., Sayers J., Croucher P. I. (2007) Investigating the interaction between osteoprotegerin and receptor activator of NF-κB or tumor necrosis factor-related apoptosis-inducing ligand: evidence for a pivotal role for osteoprotegerin in regulating two distinct pathways. J. Biol. Chem. 282, 31601–31609 [DOI] [PubMed] [Google Scholar]
- 24. Truneh A., Sharma S., Silverman C., Khandekar S., Reddy M. P., Deen K. C., McLaughlin M. M., Srinivasula S. M., Livi G. P., Marshall L. A., Alnemri E. S., Williams W. V., Doyle M. L. (2000) Temperature-sensitive differential affinity of TRAIL for its receptors: DR5 is the highest affinity receptor. J. Biol. Chem. 275, 23319–23325 [DOI] [PubMed] [Google Scholar]
- 25. Gasparian M. E., Chernyak B. V., Dolgikh D. A., Yagolovich A. V., Popova E. N., Sycheva A. M., Moshkovskii S. A., Kirpichnikov M. P. (2009) Generation of new TRAIL mutants DR5-A and DR5-B with improved selectivity to death receptor 5. Apoptosis 14, 778–787 [DOI] [PubMed] [Google Scholar]
- 26. Emery J. G., McDonnell P., Burke M. B., Deen K. C., Lyn S., Silverman C., Dul E., Appelbaum E. R., Eichman C., DiPrinzio R., Dodds R. A., James I. E., Rosenberg M., Lee J. C., Young P. R. (1998) Osteoprotegerin is a receptor for the cytotoxic ligand TRAIL. J. Biol. Chem. 273, 14363–14367 [DOI] [PubMed] [Google Scholar]
- 27. Luan X., Lu Q., Jiang Y., Zhang S., Wang Q., Yuan H., Zhao W., Wang J., Wang X. (2012) Crystal structure of human RANKL complexed with its decoy receptor osteoprotegerin. J. Immunol. 189, 245–252 [DOI] [PubMed] [Google Scholar]
- 28. Nelson C. A., Warren J. T., Wang M. W., Teitelbaum S. L., Fremont D. H. (2012) RANKL employs distinct binding modes to engage RANK and the osteoprotegerin decoy receptor. Structure. 20, 1971–1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zinonos I., Labrinidis A., Lee M., Liapis V., Hay S., Ponomarev V., Diamond P., Findlay D. M., Zannettino A. C., Evdokiou A. (2011) Anticancer efficacy of Apo2L/TRAIL is retained in the presence of high and biologically active concentrations of osteoprotegerin in vivo. J. Bone Miner. Res. 26, 630–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Picarda G., Trichet V., Téletchéa S., Heymann D., Rédini F. (2012) TRAIL receptor signaling and therapeutic option in bone tumors: the trap of the bone microenvironment. Am. J. Cancer Res. 2, 45–64 [PMC free article] [PubMed] [Google Scholar]