FIGURE 1.
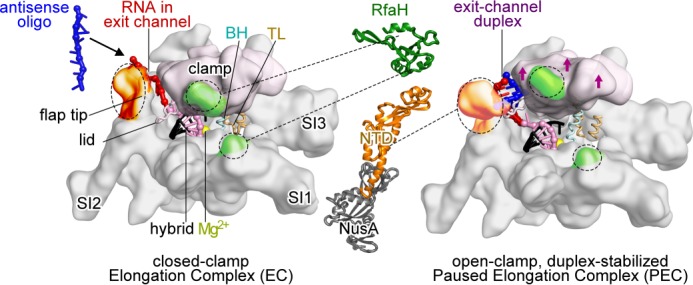
Models of E. coli RNAP EC and hairpin-stabilized paused EC. Left, closed clamp, active EC model (54) showing clamp (light pink), bridge helix (BH), trigger loop (TL), locations of major sequence insertions (SI1, SI2, and SI3) (55), flap tip, lid, active site Mg2+ (yellow), and binding sites for NusA-NTD (orange) and RfaH-NTD (green) (23, 30, 31). Most EC nucleic acids are omitted for clarity. The RNA-DNA hybrid (pink and black), exiting RNA (red), and 8-nt antisense RNA (blue) are shown. Middle, schematic representations of RfaH-NTD (Protein Data Bank code 2oug) (31, 56) and NusA-NTD, S1, KH1, and KH2 (Protein Data Bank code 1hh2) (31, 56). Dotted lines, contacts to RNAP. Right, open clamp paused EC (Protein Data Bank code 4gzy) (17). Arrows indicate clamp movement, and an 8-bp exit channel duplex (blue and red) is modeled into the expanded RNA exit channel.
