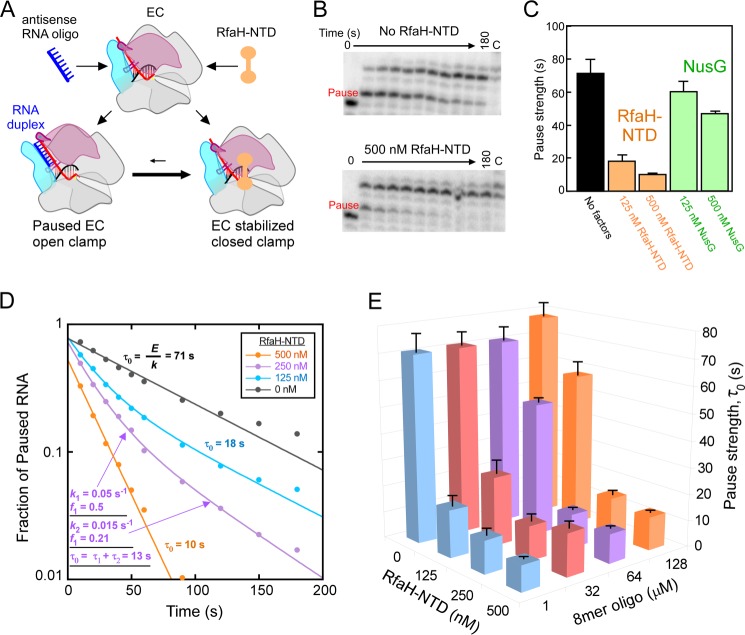
FIGURE 7.
RfaH-NTD and the 8-mer RNA oligo compete for binding to ECs. A, a model of competition between RfaH and oligo binding through the movement of the RNAP clamp domain (pink). Red, RNA; blue, antisense RNA oligo; orange, RfaH. Formation of RNA duplex requires opening of the clamp domain, which is inhibited when RfaH is bound to EC and holds the clamp in the closed position. B, representative pause assay gel panels for ECs assembled on the nucleic acid shown in Fig. 2B. ECs (C18; 50 nm) were elongated through the his pause site in the presence of 1 μm 8-mer antisense RNA oligo with or without 500 nm RfaH-NTD at 100 μm UTP and 10 μm GTP (see “Experimental Procedures”). C, pause strengths derived from the experiment shown in B and similar experiments with NusG (antisense RNA at 1 μm). Pause strengths were calculated as shown in D. D, derivation of pause strengths. The fractions of pause RNA for the experiment shown in B and similar experiments with 125 and 250 nm RfaH-NTD were plotted as a function of reaction time. Apparent fraction paused (E) and pause escape rate (k) were determined by non-linear regression. Pause strength (τ0) was calculated from E and k (τ0 = E/k). Two pause rate components were evident at 125 and 250 nm RfaH-NTD, necessitating use of a double-exponential fit. In these cases, pause strengths were calculated from the two rate components and two fractions present (τ0 = f1/k1 + f2/k2; τ0 = τ2 + τ2) as illustrated for 250 nm RfaH-NTD in the figure (see “Experimental Procedures”). E, a three-dimensional plot showing pause strengths as a function of concentrations of antisense 8-mer and RfaH-NTD present in the pause assays. Error bars, S.D.