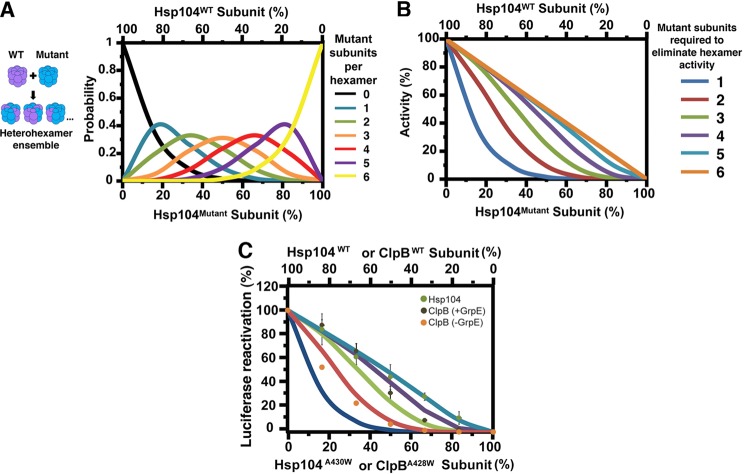
FIGURE 9.
For disordered aggregate dissolution, Hsp104 hexamers require fewer subunits able to collaborate with Hsp70 than ClpB. A, theoretical ensembles of Hsp104 hexamers containing no mutant (black), one mutant subunit (blue), two (green), three (orange), four (red), five (purple), and six mutant subunits (yellow) as a function of the percentage of mutant subunits present. See “Experimental Procedures” for more details. B, theoretical activity curves for mutant doping experiments. The activity varies as a function of the percentage of mutant subunits present assuming that hexamer activity is proportional to the number of active subunits and all subunits in a hexamer are inactive when a specified number of mutant subunits per hexamer is breached. Curves are shown for situations where one or more mutant subunits (blue), two or more mutant subunits (red), three or more mutant subunits (green), four or more mutant subunits (purple), five or more mutant subunits (light blue), or six mutant subunits (orange) are required to eliminate hexamer activity. See “Experimental Procedures” for more details. C, urea-denatured luciferase aggregates were treated with Hsp104, Hsc70, and Hdj2 plus increasing fractions of Hsp104A430W (green markers). Alternatively, luciferase aggregates were treated with ClpB, ClpBA428W, DnaK, and DnaJ. GrpE was included (gray markers) or omitted (orange markers). Where error bars are not visible, they are too small and are hidden by the marker. Luciferase reactivation (% WT activity) was then assessed. Values represent mean ± S.E. (n = 3–4). Solid lines represent theoretical activity curves for situations where one or more mutant subunits (blue), two or more mutant subunits (red), three or more mutant subunits (green), four or more mutant subunits (purple), or five or more mutant subunits (light blue) are required to eliminate hexamer activity.