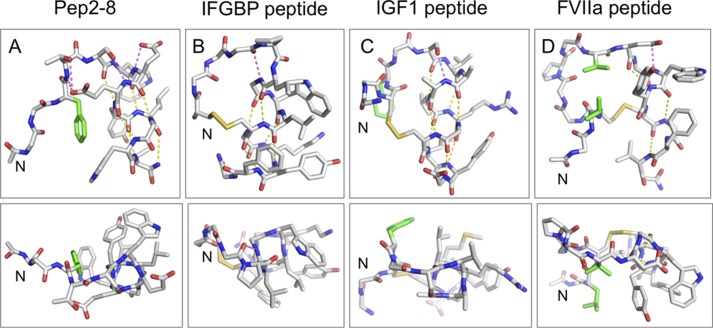
FIGURE 10.
Comparison of the Pep-2-8 fold with other strand-helix structures derived from phage display. Shown are structures of the strand-turn-helix peptides that bind to PCSK9 (Pep2-8; A), IGFBP1 (B) (39), IGF1 (C) (40), and FVIIa (D) (38). In each case, the N terminus is on the left, and the helix is on the right. The top panels show the side view of the helix, and the bottom panels show the “end on.” Side chains are depicted for those residues involved in tethering the N-terminal strand to the helix, participating in helix-cap motifs, or that form the hydrophobic surface on one face of the helix; other side chains are omitted for clarity. Helix-cap hydrogen bonds are shown in magenta, whereas helical-hydrogen bonds are in yellow. Side chains in the N-terminal strand involved in hydrophobic contacts with the helix are in green. These structures reveal a number of common features that probably contribute to the stability of the peptide fold: (i) tethering the N-terminal strand to the helix to position the helix capping motif (in the literature examples, this is achieved via a disulfide bond, whereas in Pep2-8, the hydrophobic interaction of Phe3 and hydrogen bonds from Thr4 to the side chain of Glu8 achieve the same result); (ii) helix capping motifs to help initiate the helical conformation (e.g. in Pep2-8, hydrogen bond from Ser5 side chain to the backbone amide of Glu8 and the presence of the Glu7 side chain to hydrogen-bond to its own backbone amide NH and to provide a favorable interaction with the helix dipole); (iii) predominantly hydrophobic residues along one face of the helix; the Van der Waals contacts between residues on adjacent turns help to propagate the helix and also strengthen the i, i − 4 hydrogen bonds by sequestering them from solvent. The arrangement of strand-loop-helix provides a scaffold from which to present a variety of relatively flat hydrophobic surfaces (bottom panels) for interacting with partner proteins.