Figure 5. The sequestration model for building of LMO2 protein complexes.
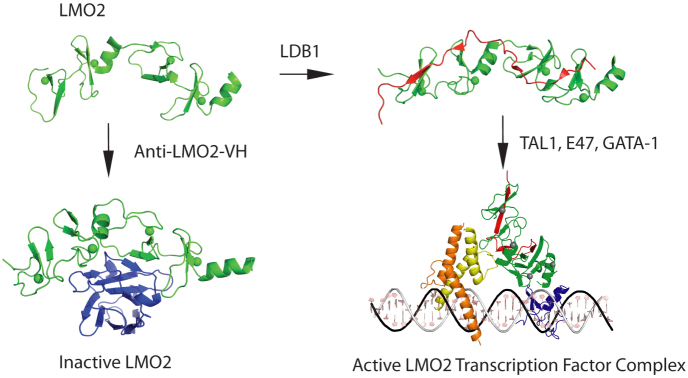
In the structural data of this paper and previous publications, it seems that the newly synthesized LMO2 protein (shown in green) is intrinsically unstable, with few structured regions. One structured region is the central short α-helix between LIM1 and LIM2 domains, giving the LMO2 protein options for interacting with partner proteins. When it binds to a natural partner such as LDB1 (shown in red), a structural constraint is imposed on LMO2 and the heterodimer can nucleate subsequent protein complex formation, depicted here the complex found in erythroid cells including GATA1. The inhibitory effect of the VH#576 (shown in blue) exploits the LMO2 hinge region and the unstable properties by sequestering LMO2 (shown in green) into a complex that has poor interacting properties with natural partners. TAL1/SCL is shown in yellow, E47 in orange and GATA1 in blue. The diagrammatic model of the pentameric complex of LMO2 (green), LDB1 (red), E47 (orange), TAL1 (yellow) and GATA-1 (blue) was generated by in silico modeling. The structure of a guide DNA including the binding motifs for E47-TAL1 (CAGGTG) and GATA-1 (GATA) transcription factors was generated using Nucleic Acid Builder of AMBER tools69. Subsequently, the structures of GATA-1, E47 and TAL1 bound DNA were derived by homology modeling70 using GATA-1 (PDB codes 1gat71 and 1gnf72) and heterodimer E47/NeuroD1 transcription factor (PDB code 2ql273) structures as templates for GATA-1 and E47-TAL1 respectively. The orientation of LMO2-LDB1 was manually adjusted by maximizing the overlap of interactions regions described in the literature: LMO2-E47-TAL136 and LMO2-GATA-174.
