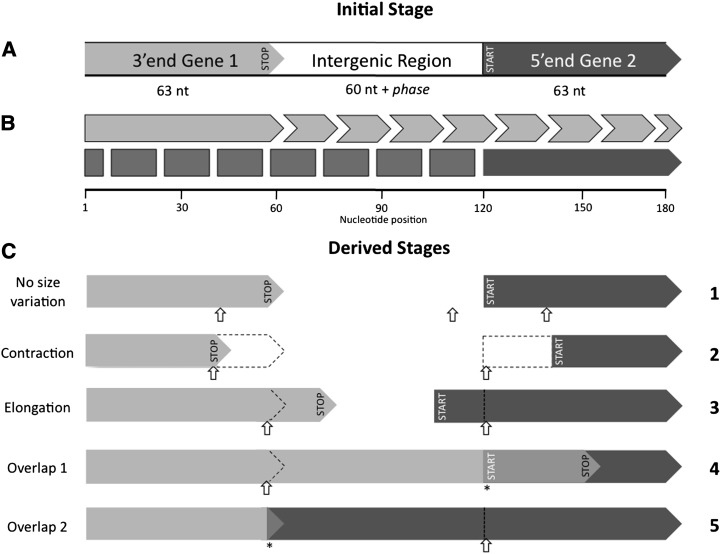
Figure 2.
Scheme of the overlapping genes formation simulations (scenario 1). (A) In the initial stage ,we generated a sequence that contained the last 21 codons of gene 1 (including the stop codon) and the first 21 codons of gene 2 (including the start codon). These two partial gene sequences have the same direction of transcription and are separated from each other by 60 plus phase nucleotides. (B) Both genes have the same limits of maximum contraction (down to one codon) or elongation (up to the end of the sequence). Light gray arrows represent the limits of gene 1 size variation, and dark gray arrows and boxes represent gene 2. (C) Next, we made random changes (small open arrows) at the nucleotide level across the sequence until an overlapping region was created (*). No gene size variation occurred if the changes occurred in the intergenic region or if they caused a synonymous/nonsynonymous mutation within the coding region (C1). However, if the changes caused the disruption of the existing start/stop codons or the creation of upstream stop codon for gene 1, then genes may alter their length (C2, C3) and ultimately create an overlap (C4, long overlap; C5, short overlap). Gene size variation was allowed to occur even without the formation of an overlapping region.