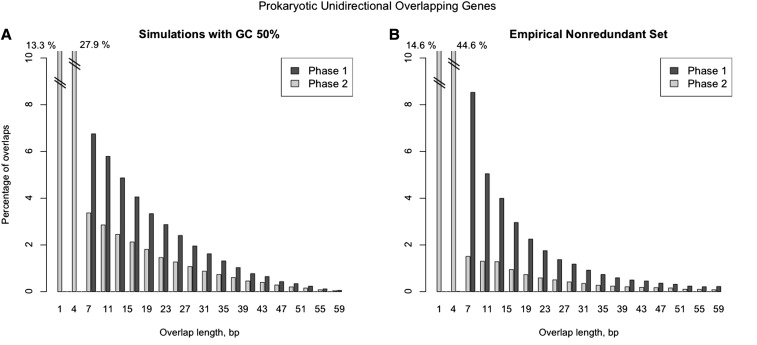
Figure 4.
Comparison of the unidirectional overlapping gene length distributions between simulated (scenario 1) and empirical data. (A) Hypothetical prokaryotic overlap lengths of unidirectional adjacent genes, calculated from simulated dataset (simulated parameters: GC content = 50%; criterion = Both; proportions of start codons = TRUE). Frequencies of overlaps in both phases were weighted according to the mutation rate between phase 1 and phase 2 simulations (rate = 1.59; see Table 1). Frequency proportions used were as follows: phase 1 = 38.6%; and phase 2 = 61.4%. (B) Distribution of empirical prokaryotic overlap lengths of unidirectional adjacent genes calculated from a nonredundant set of 400,363 overlapping gene pairs (phase 1 = 31.6%; phase 2 = 68.4%).