Abstract
Background
Non-syndromic hearing loss (NSHL) is the most common sensory impairment in humans. Until recently its extreme genetic heterogeneity precluded comprehensive genetic testing. Using a platform that couples targeted genomic enrichment (TGE) and massively parallel sequencing (MPS) to sequence all exons of all genes implicated in NSHL, we test 100 persons with presumed genetic NSHL and in so doing establish sequencing requirements for maximum sensitivity and define MPS quality score metrics that obviate Sanger validation of variants.
Methods
We examined DNA from 100 sequentially collected probands with presumed genetic NSHL without exclusions due to inheritance, previous genetic testing, or type of hearing loss. We performed TGE using post-capture multiplexing in variable pool sizes followed by Illumina sequencing. We developed a local Galaxy installation on a high performance-computing cluster for bioinformatics analysis.
Results
To obtain maximum variant sensitivity with this platform 3.2–6.3 million total mapped sequencing reads per sample are required. Quality score analysis showed that Sanger validation is not required for 95% of variants. Our overall diagnostic rate was 42% but varied by clinical features from 0% for persons with asymmetric hearing loss to 56% for persons with bilateral autosomal recessive NSHL.
Conclusions
These findings will direct the use of TGE and MPS strategies for genetic diagnosis for NSHL. Our diagnostic rate highlights the need for further research on genetic deafness focused on novel gene identification and an improved understanding of the role of non-exonic mutations. The unsolved families we have identified provide a valuable resource to address these areas.
Keywords: Deafness, hearing loss, targeted genomic enrichment, sequence capture, massively parallel sequencing
INTRODUCTION
Hearing loss is the most common sensory deficit in humans. It is diagnosed in 1 of every 500 children in North America and Europe [1]; it impacts one-third of persons at least 65 years of age and in aggregate affects 360 million people worldwide (WHO data, http://www.who.int/pbd/deafness/estimates/en/index.html). Both environmental and genetic factors are known to damage hearing. Examples of the former are noise, ototoxic drugs, and viral and bacterial infections; amongst the latter, a staggering number of genes – 67– and genetic variants – 990 – have been implicated in inherited hearing loss.
Hearing loss that occurs in the absence of any other abnormal physical findings is referred to as non-syndromic hearing loss (NSHL). The extreme genetic heterogeneity of NSHL has made comprehensive genetic diagnosis based on Sanger sequencing impractical. To address the daunting heterogeneity and minimize the labor and expense of gene-by-gene Sanger sequencing, various strategies have been adopted to prioritize genes for mutation screening. Amongst these strategies are segregation analysis and the use of phenotypic data to predict genotypes. Whilst these approaches have been variably successful when applied to large dominant families or individuals with unique audioprofiles, for most families the likely genetic cause of deafness remained difficult to establish and made NSHL a diagnosis of exclusion.
To address the genetic heterogeneity of deafness and minimize the labor and expense of gene-by-gene Sanger sequencing, we developed a platform (referred to as OtoSCOPE®) that couples targeted genomic enrichment (TGE) and massively parallel sequencing (MPS) to capture and sequence all exons of all genes implicated in NSHL. In a proof-of-principle study we showed that OtoSCOPE® is sufficiently sensitive and specific for clinical diagnostics [2]. There have been several other recent studies using enrichment and MPS technologies with similar positive results [3–5].
Our goal in this study was to show that comprehensive genetic testing for NSHL should be routine in the clinical evaluation of the deaf and hard-of-hearing person. To that end, we examined levels of coverage and sequencing reads to determine the maximum sensitivity of our TGE/MPS platform and to define quality score levels that make Sanger validation of variants unnecessary. We illustrate the power of TGE + MPS as applied to 100 patients with presumed genetic hearing loss. Although we sequenced only a small proportion of the genome, we uncovered a large degree of genetic variation and so developed a standardized prioritization scheme for variant interpretation.
MATERIALS AND METHODS
Subjects
Subjects were sequentially accrued probands referred to our laboratory as part of a large study of genetic hearing loss and met the following broad criteria for genetic NSHL: 1) no apparent syndromic features and a family history of hearing loss, or 2) if an isolated proband, a thorough clinical evaluation to rule out obvious causes of environmental hearing loss. We characterized patients based on inheritance as: 1) presumed autosomal recessive (consanguineous or affected siblings or both), 2) autosomal dominant, and 3) sporadic (no family history of hearing loss). Subjects were not excluded based on previous genetic testing, age of onset, inheritance, habilitation used, or type of hearing loss. Although clinical evaluation varied based on referral, in general we required the following clinical data for enrollment: 1) audiogram and/or auditory brainstem response (ABR), 2) complete physical exam and history, 3) detailed family history, and in some cases 4) computed tomography (CT) of the temporal bones and/or fundoscopy. We defined hearing loss severity as: mild, 21–40 dB; moderate, 41–70 dB; severe, 71–95 dB; and profound, >95 dB. Data from six samples (A5 – A10) have been previously published but were reanalyzed using the methods described below [2]. All methods were approved by the IRB at the University of Iowa.
Library Preparation, Targeted Genomic Enrichment, and Sequencing
We developed a platform (OtoSCOPE®) for sequencing all exonic regions of all known deafness genes using four successive implementations of TGE + MPS (Supplementary Tables 1 and 2) [2]. Library preparation was performed either manually or using liquid-handling automation equipment. For samples prepared manually, we used a modified solution-phase targeted genomic enrichment protocol to maximize sample retention and minimize DNA input [6]. For samples prepared using automation equipment we used the manufacturer’s recommended protocol (Bravo System, Agilent Technologies, Santa Clara, CA, USA) as described [7]. In brief, genomic DNA was assessed for quality by spectrophotometer (260/280 = 1.8–2.0) and gel electrophoresis (to ensure high molecular weight gDNA), and quantitated using the Qubit system. 1–3 micrograms of DNA was randomly fragmented using focused acoustics (Covaris Inc., Woburn, MA, USA), ends were repaired, A-tails were added, and sequencing adaptors were ligated prior to the first amplification. Solid-phase reverse immobilization (SPRI) purifications were performed between each enzymatic reaction. Hybridization and capture with RNA baits was followed by a second amplification prior to pooling for sequencing. In all cases, a minimum number of amplifications possible was used (typically 8 cycles for the pre-hybridization PCR (range 8–10 cycles using NEB Phusion HF Master Mix) and 14 cycles for the post-hybridization PCR (range 12–16 cycles using Agilent Herculase II Fusion DNA Polymerase)). Cycling parameters are detailed in the previously published method [6].
We performed sequencing using the Illumina GAIIx, Illumina MiSeq, or Illumina HiSeq using 100 bp paired-end reads in all cases. All samples, excluding set 1, were barcoded and multiplexed prior to sequencing. We used post-capture multiplexing in all cases. Candidate variants were confirmed via Sanger sequencing using custom primers – all primer sequences are available on request.
Bioinformatics
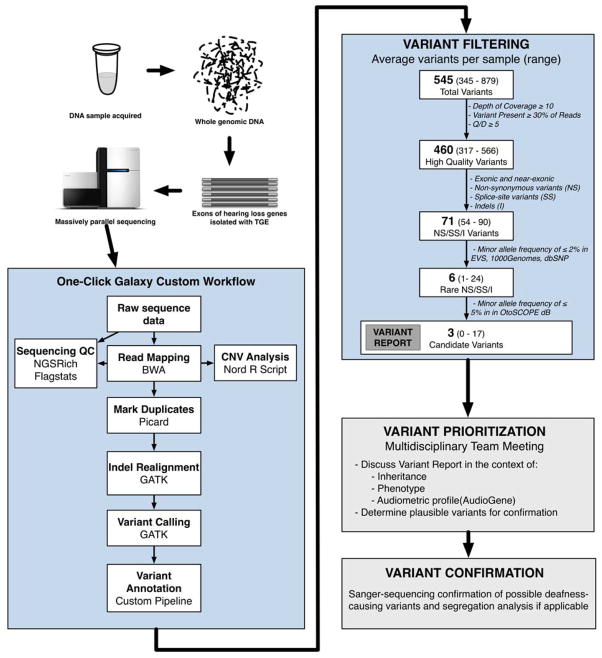
We implemented a local installation of the open-source Galaxy software running on a high-performance computing cluster at the University of Iowa for bioinformatics analysis (overview is shown in Figure 1). There is a dedicated compute queue for Galaxy as well as shared compute resources that can be leveraged for minimized job wait time and continuous availability. A distributed file system, Glustre (http://lustre.org), serves as the storage platform for the datasets that are computed and imported into the Galaxy portal; the Glustre storage servers are accessed over an infiniband network. The computing environment was designed to be fault tolerant with job failures due to hosting issues automatically re-launched out to cluster without intervention from the end user.
Figure 1.
Overview of Bioinformatics and Variant Filtering for Diagnostics with OtoSCOPE®.
We used a combination of custom and publicly available tools to develop our bioinformatics pipeline running in Galaxy. Read mapping was performed with Burrows-Wheeler Alignment (BWA, [8]), duplicate removal with Picard, local re-alignment and variant calling with GATK [9], enrichment statistics with NGSRich [10], and variant annotation with a custom tool.
We used two primary measures to classify variant quality: 1) %Obs, n observed reads/n total reads at variant position, 2) QD, Phred-like Quality/Depth at variant position. We incorporated data from the Exome Sequencing Project, a publicly available resource of exome sequencing data from more than 6,500 samples comprising two populations, European Americans (EA) and African-Americans (AA) (http://evs.gs.washington.edu/EVS/, data accessed 9/2012), 1000 Genomes project (http://1000genomes.org, data accessed 9/2012), and dbSNP137 (http://www.ncbi.nlm.nih.gov/projects/SNP/). Pathogenicity prediction was performed using all scores available in dbNSFP version 2 [11].
During variant prioritization, we used the following three definitions: 1) high quality variants, %Obs ≥ 30% and Q/D ≥ 5; 2) rare NS/SS/I (Non-Synonymous, Splice Site, Indel) variants present at Minor Allele Frequences (MAFs) ≤0.005 for recessive/sporadic cases, excluding GJB2, or ≤0.0005 in dominant cases in population-scale databases including dbSNP, 1000 Genomes and EVS; and 3) candidate variants, present with a frequency of < 0.05 in all other samples run on this platform as a method to rule out platform-specific errors.
The result of the Galaxy analysis pipeline is a PDF report (Supplemental File 1) depicting the quality of the sequence data as compared to expected results and variant results. The report provides a dynamic visual alert of sequence/mapping quality and variant quality/likelihood, automates the identification and interpretation of strong potential variants, and generates an alert when data quality is suspect and a sample re-run is needed.
Copy number variants (CNVs) were determined using a published method that normalizes sequencing depth among samples, identifies outliers, and calls via a sliding-window method [12]. We validated this technique for the STRC region (15q15.3), which is a common site for CNVs in the general population and in persons with mild-to-moderate deafness [13] using an adapted MLPA probeset [14] in 64 GJB2-negative probands and blinded control individuals. We found 100% concordance between the MLPA and bioinformatics methods: we identified 5 homozygous and 4 heterozygous deletions of the STRC region. We then used this bioinformatics method to analyze all targeted genes for CNVs.
RESULTS
Sequencing and Coverage Results
We performed TGE+MPS on 100 DNA samples from persons with presumed genetic deafness in six different sets with different combinations of methods for library preparation and sequencing, as shown in Table 1. No significant differences in diagnostic rates, described below, were identified between sets (ANOVA, p = 0.096). In all cases, both the average depth-of-coverage of targeted regions (average 1,400X) and the percent of targeted bases covered at our variant calling threshold of 10 sequencing reads were high (average 97.8%).
Table 1.
Sequencing Statistics for 100 Samples from Patients with Apparent Genetic Deafness. DOC, depth of coverage
| Sample Set | n Samples | OtoSCOPE version (n genes) | Library Prep Method | Sequencing Platform | Multiplexing (n/pool) | Avg Total Reads (range) | Avg % Reads Mapped (range) | Avg % Capture Efficiency (range) | Avg DOC (range) | Avg Target Covered ≥ 1X | Avg Target Covered ≥ 10X | Avg Target Covered ≥ 20X |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 7 | 1 (54) | manual | GAIIx | - | 39,730,574 (3,594,022–66,352,788) | 96.3% (91.3–97.9%) | 17% (10–28%) | 1,060 (191–2,385) | 99.8% (99.7–99.9%) | 99.4% (98.8–99.7% | 99.0% (96.7–99.6%) |
| 2 | 7 | 2 (59) | manual | GAIIx | 10 | 52,224,268 (41,037,728–68,215,072) | 51.0% (40.1–60.1%) | 27% (18–38%) | 2,944 (1,423–5,118) | 99.9% (99.8–99.9%) | 99.7% (99.5–99.7%) | 99.5% (99.5–99.7%) |
| 3 | 20 | 3 (66) | manual | HiSeq | 4, 10, or 12 | 26,423,265 (9,867,026–63,482,934) | 93.3% (82.0–98.3%) | 56% (47%–66%) | 2,740 (1,094–5,417) | 99.0% (97.8–99.9%) | 98.3% (96.4–99.6%) | 97.9% (95.6–99.4%) |
| 4 | 39 | 4 (66) | automated | HiSeq | 96 | 2,720,037 (1,282,442–4,909,916) | 97.0% (95.5–97.5%) | 62% (43–68%) | 247 (120–486) | 99.4% (98.9–99.6%) | 97.3% (93.7–98.6%) | 93.6% (84.9–97.3%) |
| 5 | 15 | 4 (66) | manual | HiSeq | 12 | 30,056,966 (9,583,798–45,361,948) | 98.3% (95.1–98.9%) | 59% (31–65%) | 2,980 (1,060–4,107) | 99.8% (99.7–99.8%) | 99.5% (99.2–99.6%) | 99.3% (98.8–99.5%) |
| 6 | 12 | 4 (66) | manual | MiSeq | 6 | 1,477,023 (690,352–2,993,118) | 75.2% (62.33–89.3%) | 41% (32–54%) | 160 (62–327) | 99.2% (98.8–99.5%) | 94.6% (90.5%–98.0%) | 77.6% (62.3–92.0%) |
|
| ||||||||||||
| Average (Stdev) | 100 | - | - | - | - | 17,468,094 (18,490,901) | 90.6% (13.7%) | 52.4% (15.4%) | 1,400 (1,443) | 99.4% (0.05%) | 97.8% (1.9%) | 95.1% (5.4%) |
For in-depth analysis of coverage statistics, we used data solely from a single version of the OtoSCOPE® platform, v4 (n=67 samples), because comparison between different targeted genomic regions would prohibit a normalized comparison. We found the number of mapped sequencing reads correlated logarithmically with 10X depth of coverage (r2 = 0.6119) such that 95%, 98%, or 99% coverage of targeted bases at 10X depth required 566,935, 6,249,425 and 13,908,351 mapped sequencing reads, respectively (Supplementary Figure 1).
While base coverage is important for sensitivity, we also aimed to ensure that all possible variants within the targeted regions were identified. To address this issue, we divided data into a high coverage data set (n = 17, average depth of coverage = 3,185X) and a low coverage data set (n = 50, average depth of coverage = 227X). In the low coverage data set, we identified a significantly lower number of high quality variants (p = 0.004) and rare NS/SS/I variants (defined in Methods; p = 0.023), although there was no significant difference in the number of common NS/SS/I variants (defined in Methods, p = 0.069) or candidate variants (defined in Methods; p = 0.478). As shown in Supplementary Figure 2, the low coverage dataset showed a linear correlation between increasing depth-of-coverage and variant identification for high quality and common NS/SS/I variants (R = 0.200, 0.156, respectively) but not for rare NS/SS/I variants (R= 0.043). In the high coverage data set, in contrast, variant detection was not a function of depth of coverage (R > 0.010 in all cases).
The average number of high quality and common NS/SS/I variants from the high coverage data set was 510 and 75 variants, respectively. In order to obtain these numbers of variants, the average depth-of-coverage must be 341X or 311X. We chose 341X (the most conservative level) as a quality control metric. When depth-of-coverage was plotted against mapped reads, there is a strong linear relationship, R = 0.965 (Supplementary Figure 3). 341X sequence depth is obtained when a sample has 3,225,290 mapped sequencing reads. Thus, for coverage of targeted regions at 98%, 6,249,425 mapped reads are required, but for maximum variant identification sensitivity, at least 3,225,290 mapped reads are required per sample. These values can be used as quality control metrics going forward to ensure that samples are sequenced adequately for clinical testing.
Variant Quality Analysis
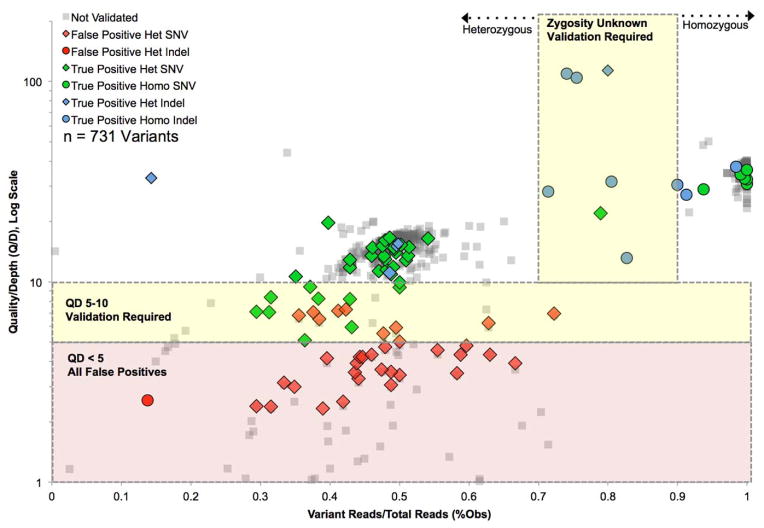
Next, we aimed to determine the necessity for Sanger validation by correlating quality scores from MPS data with Sanger sequencing results. We stratified MPS data using two metrics of variant quality, as shown in Figure 2: variant percent observed (the number of variant reads divided by depth of coverage at the variant position) and QD (Phred-like Quality score divided by Depth). We ascertained all non-synonymous or indel exonic variants for 12 samples chosen at random from this dataset and then we chose a distribution of variants falling within varying levels of quality to validate with Sanger sequencing (Figure 2). In these samples, we identified 692 exonic variants including 661 SNVs (95.5%) and 31 indels (4.5%). Of these, we Sanger validated 54. We combined these data with the 39 causative mutations Sanger validated (described below, 31 SNVs and 8 indels) to arrive at 93 Sanger sequenced variants: 75 SNVs and 16 indels out of a total of 731 variants.
Figure 2.
Validation Of MPS Variants At Varying Levels Of Quality Establish Cut-Offs For Required Sanger Sequencing Validation. 731 total variants in 12 samples are shown. Yellow shading indicates validation required due to high false positive rate (QD 5–10) or indeterminate zygosity status (%Obs 70–90%). Red shading indicates all false positives. Overall, validation is required for 4.8% of 731 variants.
There are two purposes for Sanger sequencing validation – confirmation of the variant (to rule out false-positives) and confirmation of zygosity status. As shown in Figure 2, 609 of the 731 variants (83.3%) had a QD ≥10; 100% of the 44 variants we validated above this level were true positives. 26 variants (3.6%) had a QD between 5 and 10, and of 20 we Sanger sequenced, only 10 (50%) were true positives. 96 of the 731 variants (13.1%) fell below a QD of 5 and of the 27 we Sanger sequenced, all were false positives. Therefore, Sanger sequencing should be performed to validate variants with a QD greater than 5 but less than 10; variants with a QD above 10 do not need to be Sanger validated and variants with a QD below 5 are false positives and should not be considered. We did not identify a single unifying reason for low QD values (such as “difficult” genes or samples) for low QD values.
With respect to zygosity, generally in MPS studies zygosity is determined by %Obs, such that heterozygous variants are >30% and <90% Obs, and >90% Obs is considered homozygous. In this study we did not filter by %Obs but considered all variants, validating the subset described above and shown in Figure 2. We found that when %Obs was between 70–90% Obs for simple transitions and transversions (9 of 731 variants, 1.2%), the zygosity status was indeterminate. For indels, zygosity was more indeterminate even when QD was above 10 (including one true positive indel at 14%). Based on these data we recommend validation of zygosity status for all variants falling between 70–90% Obs.
In total, of all 731 variants we examined, using these guidelines for validation to rule out false positives and to determine zygosity status, Sanger sequencing confirmation would be required for 35 variants, or 4.8%.
Variant identification and diagnostic rates in deaf patients
To facilitate high-throughput bioinformatics analysis with minimal user intervention, we developed a robust and easily implemented workflow using an open-source Galaxy framework with automated filtering and report generation (see Figure 1 and Methods). We performed comprehensive genetic testing for deafness using OtoSCOPE® on 100 sequentially acquired DNA samples from patients with presumed genetic deafness (Table 2), which we classified as autosomal recessive (39%), autosomal dominant (29%) or sporadic (32%). By age-of-onset, 53% of patients had congenital deafness, and by severity, in 49% the hearing loss was severe-to-profound. Genetic testing, usually for variants in GJB2, had been completed in 29% of patients.
Table 2.
Clinical Characteristics for 100 Individuals with Apparent Genetic Deafness.
| Demographic | Category | n (%) |
|---|---|---|
| Sex | Male | 44 (45%) |
| Female | 54 (55%) | |
|
| ||
| Inheritance | Presumed Autosomal Recessive | 39 (39%) |
| Autosomal Dominant | 29 (29%) | |
| Sporadic | 32 (32%) | |
|
| ||
| Age Of Onset | Congenital | 53 (53%) |
| Childhood (<18 Years) | 34 (34%) | |
| Adult (>18 Years) | 9 (9%) | |
|
| ||
| Type Of Hearing Loss | Mild-Moderate | 42 (43%) |
| Severe-Profound | 49 (50%) | |
| Asymmetric | 7 (7%) | |
|
| ||
| Previous Testing | No Testing | 71 (71%) |
| GJB2/GJB6 Testing | 19 (19%) | |
| Other Gene Testing | 6 (6%) | |
| GJB2/GJB6 and Another Gene Testing | 4 (4%) | |
Using our custom analysis pipeline (Figure 1 and Methods), we identified on average 545 variants per patient, including 460 high quality variants (%Obs ≥ 30% and QD ≥ 5), 71 NS/SS/I variants, 6 rare NS/SS/I variants, and 3 candidate variants. By interpreting and classifying these variants on a case-by-case basis at an interdisciplinary meeting in the context of all available clinical information, we were able to provide a definitive diagnosis of genetic deafness in 42 patients (70 causative mutations in 21 genes; Supplementary Data File 1). All causative variants in this study were validated by Sanger sequencing in the proband and other members of the family, if segregation analysis was possible.
As shown in Table 3, we evaluated our diagnostic rate (ability to identify a causative mutation) by clinical characteristics including inheritance, previous genetic testing, hearing loss onset, and type of loss. Diagnostic rates varied from as low as 0% (patients with asymmetric hearing loss) to 56% for cases of presumed recessive deafness. There were no significant differences between these groups, although inheritance trended towards significance (p = 0.063, test for equality of proportions without continuity correction).
Table 3.
Diagnostic Rates Based on Clinical Characteristics for 100 Individuals with Apparent Genetic Deafness.
| Category | % Diagnosis Provided (n) | Total n | |
|---|---|---|---|
| All Cases | - | 42% (42) | 100 |
|
| |||
| Inheritance | Presumed Recessive | 56% (22) | 39 |
| Dominant | 31% (9) | 29 | |
| Sporadic | 34% (11) | 32 | |
|
| |||
| Previous Testing | No testing | 41% (29) | 71 |
| DFNB1 testing | 53% (10) | 19 | |
| Other gene test (non-DFNB1) | 17% (1) | 6 | |
| DFNB1 and other gene test | 50% (2) | 4 | |
|
| |||
| Hearing Loss Onset | Unknown Onset | 50% (2) | 4 |
| Congenital | 49% (26) | 53 | |
| Childhood | 35% (12) | 34 | |
| Adult Onset | 22% (2) | 9 | |
|
| |||
| Type Of Loss | Unknown Type | 50% (1) | 2 |
| Mild-Moderate | 45% (19) | 42 | |
| Severe-Profound | 45% (22) | 49 | |
| Asymmetric | 0% (0) | 7 | |
As shown in Table 4, the majority of causative mutations identified were SNVs (44, 61%), followed by indels (19, 26%) and large deletions (9, 13%). Although 23% of patients had earlier GJB2 mutation screening and were GJB2-negative, we identified GJB2 as the causative gene in 6 cases. In 1 of these cases, the person had been previously screened for mutations in GJB2 by allele-specific PCR, which could not have detected the causative mutation identified by OtoSCOPE®.
Table 4.
Causative Mutations Identified in 100 Individuals with Presumed Genetic Deafness.
| Causative Mutation By Gene | Gene | n Patients | % of all Solved Patients |
|---|---|---|---|
| GJB2 | 6 | 14.3% | |
| STRC | 4 | 9.5% | |
| CDH23 | 3 | 7.1% | |
| MYH14 | 3 | 7.1% | |
| MYO15A | 3 | 7.1% | |
| MYO7A | 3 | 7.1% | |
| SLC26A4 | 3 | 7.1% | |
| MITOCHONDRIAL | 2 | 4.8% | |
| TECTA | 2 | 4.8% | |
| WFS1 | 2 | 4.8% | |
| ACTG1, COCH, DIAPH1, EYA4, GPR98, KCNQ4, MYH9, MYO6, OTOA, USH1G, USH2A | 1 each | 2.4% each | |
|
| |||
| Total | 42 | 100% | |
| Causative Mutation By Type | Type | n Mutations | % of all Causative Mutations |
|---|---|---|---|
| Single Nucleotide Variant | 44 | 62.9% | |
| Indel | 17 | 24.3% | |
| Large Deletion | 9 | 12.9% | |
|
| |||
| Total | 70 | 100% | |
Of 26 patients with congenital severe-to-profound deafness who had not had genetic testing, 14 (54%) were solved, and of this number, 5 (36%) had GJB2-related hearing loss (Table 4). Mutations in STRC were second most common overall (4 patients, 9.3%) and the most common cause of mild-to-moderate downing-sloping hearing loss (4 of 18 solved, 22.2%). However, when mutations in the Usher syndrome genes are taken as a single group, this diagnosis was most frequent (10 patients, 23.2%).
Secondary findings included 7 carriers of Usher syndrome mutations, 7 carriers of variants causing Wolfram Syndrome (mutations in the gene WFS1), 1 carrier of a Pendred syndrome mutation, 1 carrier of a mutation in COL11A2 reported to cause Stickler syndrome, and 1 carrier of a mutation in GJB2 (Supplemental Data File 1). Thus of 100 patients, 17 had secondary findings of clinical relevance. This number includes 6 persons in whom we identified causative NSHL mutations in other genes, but who were also carriers of other clinically relevant mutations that caused NSHL, Usher syndrome or Wolfram Syndrome.
DISCUSSION
In this study we sought to show that comprehensive genetic testing for NSHL can now be routine in the clinical evaluation of the deaf and hard-of-hearing person. To facilitate testing, we developed a TGE+MPS framework, improving the wet-lab methodology, analysis, and validation. We then used this framework to screen a large number of deaf probands. Our results support the following conclusions: 1) Stringent and platform-specific quality measures can make Sanger validation unnecessary; 2) In approximately half of persons with presumed hereditary hearing loss, it is possible to identify a genetic cause using TGE+MPS technology; and 3) The ‘unsolved’ rate for genetic deafness underscores the extreme heterogeneity of deafness and highlights the need for continued research.
Clinical Utility and Diagnostic Rates
In developing TSE+MPS, our goal was to provide a personal genomic test that was clinically useful, improved the care of deaf and hard-of-hearing persons, and reduced over-all healthcare expenditure by making other diagnostic tests unnecessary. To our knowledge, this study is the first to test the utility of comprehensive genetic testing on a large number of patients with presumed genetic deafness. Our diagnostic rate – that is our ability to identify a genetic cause for hearing loss – varied based on clinical features such as type of inheritance and degree of hearing loss. For example, for persons segregating dominant hearing loss, our diagnostic rate of 31% can be partially explained by the fact that of the 47 reported ADNSHL loci, genes have been identified for only 27 (57%). In contrast, our diagnostic rate for ARNSHL was 56% and 40 of 55 (73%) genes at ARNSHL loci are known.
Our data highlight the importance of two clinical findings that are strongly indicative of genetic hearing loss: family history and symmetry of hearing loss. In patients with sporadic deafness for example, the diagnostic rate was 34% as compared to 56% for recessive cases; the diagnostic rate for persons with asymmetric hearing loss was 0% compared to 45% for persons with bilaterally symmetric hearing loss. We therefore recommend restricting genetic testing to persons with a positive family history of hearing loss or in sporadic cases, to persons with symmetric hearing loss.
Of the 127,231 reported disease-causing mutations in the Human Gene Mutation Database (HGMD.org), 112,408 (88.3%) are NS/SS/I variants. Extrapolating these data to the OtoSCOPE® diagnostic rate would mean that even when ALL deafness-causing genes are screened, since only coding variants and exonic CNVs are currently considered, the theoretic maximum diagnostic rate should be ~88%. Improving this rate will require a better understanding of gene- and inner ear- specific promoters and enhancers and the effect of non-coding variation on the genome in general to provide more comprehensive screening. In order to improve our ability to interpret genetic variants we have developed a freely available database, the Deafness Variation Database (DVD, http://deafnessvariationdatabase.org) for dissemination of information on all variants encountered during our screening. The DVD will be routinely updated with our interpretation of variants as
Sanger Sequencing Validation
Clinical genetic testing is based on Sanger sequence identification of the variant in question. As TGE+MPS platforms have been developed, Sanger validation of variants has been incorporated into the diagnostic pipeline. However this step is redundant when errors are removed by stringent mapping and realignment, as well as quality-score recalibration. To define high-standard metrics we examined all non-synonymous and indel variants for 12 samples as well as all causative mutations that were Sanger validated (731 variants) (Figure 1). All variants with a QD ≥ 10 were true positives and all variants with a QD <5 were false positives. Zygosity status of indels was difficult to determine, especially between 70–90%.
Based on these data, using our TGE+MPS platform there is no need to validate SNVs with a QD ≥ 10 or < 5, however if the QD is between these values or the variant is an indel, Sanger validation must be done. In addition, zygosity status must be validated when the %Obs is between 70–90%. These metrics mean that Sanger sequencing is required for only about 5% of discovered variants.
We believe that a comparable analysis could be applied to other diagnostic platforms that use Illumina sequencing and a similar analysis pipeline. However, we hypothesize that differences in library preparation, sequencing type used, and analysis method could alter the quality scores. Therefore, we suggest that each laboratory wishing to implement a similar TGE+MPS platform should perform a rigorous quality analysis prior to considering foregoing Sanger validation. Finally, the issue of sample mix-up is relevant and Sanger sequencing can be a valuable method for sample quality assurance, however each laboratory should develop standard operating protocols that use an orthologous method for sample quality assurance and this need not be Sanger sequencing.
Exome versus OtoSCOPE®
As an alternative to a limited, disease-specific platform like OtoSCOPE®, it is possible to sequence the exome and filter it selectively, interrogating only those genes that are known to cause hearing loss. This approach offers the possibility of ‘sequencing once’ and ‘interrogating multiple times’, as more deafness-causing genes are identified. To test the feasibility of this strategy, we compared 12 exomes sequenced using SureSelect 51 Mb, Illumina sequencing to OtoSCOPE® samples and found that while overall exome coverage was good (average 92.3%, stdev 3.5%, at 10X depth of coverage), coverage of OtoSCOPE®-specific regions was poor (average 70.8%, stdev 5.5%, at 10X depth of coverage). However, when only coding base-pairs of OtoSCOPE® were considered, exome coverage was better (average 94.8%, stdev 3.0%, at 10X depth of coverage), although it was still lower than OtoSCOPE® coverage (average 98.2%, stdev 1.3% at 10X depth of coverage for all regions targeted). We believe this difference, which other groups have also reported, reflects omission of many organ-specific isoforms in the standard exome target regions [15].
There are two additional considerations that currently favor the use of a limited TGE platform: 1) the cost and ease of enrichment, sequencing and analysis; and, 2) the ethical issues associated with secondary findings. Exome sequencing, for example, generates nearly seven times as many reads as our more limited platform (118 million reads per sample versus 17 million reads per sample). In addition, exome file sizes are 30X larger and take 10X longer to analyze. Secondary findings (variants known to be associated with a clinical phenotype other that the phenotype under investigation) are also germane and were present in 17 of 100 patient samples we screened using OtoSCOPE® including 6 persons in whom we determined causative mutations but who also carried unrelated but clinically relevant mutations. In contrast, in any exome there will be hundreds of variants of potential clinical importance that need to be considered individually [16,17]. Identification of these variants will affect genetic counseling significantly, as patients will need to be counseled about mutations in genes causing the phenotype for which they sought the test and also for clinically relevant mutations found in other genes. The amount of time and effort spent on addressing secondary findings is an area of on-going research and investigation.
Newborn Hearing Screening
Early detection of deafness through newborn hearing screening (NBHS) programs has positively affected childhood developmental outcomes. However there are several limitations to the current identification method: (1) the first screening method – otoacoustic-emissions – measures outer hair cell response, but has a high false-positive rate and misses specific NSHL phenotypes; (2) as a newborn screen, early-onset or late-onset (non-congenital) deafness is missed; (3) secondary screening is routinely required; and (4) there is no focus on differentiating environmental versus genetic causes of hearing loss, which would aid in family counseling [18,19]. Molecular genetic testing addresses each of these weaknesses by providing a specific diagnosis not reliant on phenotype and not requiring confirmatory follow-up testing. One long-term goal in the field of genetic hearing loss has been to develop a comprehensive genetic testing platform that can quickly and efficiently provide a genetic diagnosis for newborns. We can imagine in the near future that genetic testing for hearing loss will complement or replace the phenotypic-based NBHS.
Supplementary Material
Figure S1. Mapped Sequencing Reads Correlate With 10X Average Depth Of Coverage. 11 outlying samples removed with > 10 Stdev coverage from the mean at 1X, 10X, and 20X.
Figure S2. Variant Discovery is Dependent on a Saturating Depth Of Coverage. Each point represents a single sample data set. Inset shows low coverage data set.
Figure S3. Mapped Sequencing Reads Correlate Linearly With Average Depth of Coverage.
Table S1. General Overview of OtoSCOPE® Platform by Version.
Table S2. Genes Included on OtoSCOPE®.
Acknowledgments
This work was supported in part by NIDCD RO1s DC003544, DC002842 and DC012049 to RJHS, as well as NIDCD 1F30DC011674 to AES, and an NHMRC Overseas Biomedical Postdoctoral Training Fellowship (ID 546493) to MSH
Footnotes
COMPETING INTERESTS
SJ, HR, ACG, BN, SH, and EML are employees of Agilent Technologies Inc., which manufactures and offers for commercial sale targeted genomic enrichment kits like the one used in this work. All other others have no competing financial interests.
AUTHORS’ CONTRIBUTIONS
Designed study: AES, MSH, RWE, EML, RJHS; Performed Experiments: AES, MSH, RWE, HR, SJ, ACG, CMS, SDH; Performed analysis: AES, EAB-Z, APD, KRT, RJHS; Collected and assembled data: AES, EAB-Z, SH, BN; Contributed tools: EAB-Z, BN, APD, KRT, TES, TAB, TLC, EML; Contributed samples and/or reagents: HR, SJ, SH, BK, EML; Drafted manuscript: AES, RJHS; All authors contributed to, edited and reviewed the final manuscript.
References
- 1.Morton N. Genetic epidemiology of hearing impairment. Ann N Y Acad Sci. 1991;630(1):16–31. doi: 10.1111/j.1749-6632.1991.tb19572.x. [DOI] [PubMed] [Google Scholar]
- 2.Shearer AE, DeLuca AP, Hildebrand MS, Taylor KR, Gurrola J, Scherer S, Scheetz TE, Smith RJH. Comprehensive genetic testing for hereditary hearing loss using massively parallel sequencing. Proc Natl Acad Sci U S A. 2010;107(49):21104–21109. doi: 10.1073/pnas.1012989107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tang W, Qian D, Ahmad S, Mattox D, Todd NW, Han H, Huang S, Li Y, Wang Y, Li H, Lin X. A Low-Cost Exon Capture Method Suitable for Large-Scale Screening of Genetic Deafness by the Massively-Parallel Sequencing Approach. Genetic Testing and Molecular Biomarkers. 2012 doi: 10.1089/gtmb.2011.0187. 120405132347004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schrauwen I, Sommen M, Corneveaux JJ, Reiman RA, Hackett NJ, Claes C, Claes K, Bitner-Glindzicz M, Couck P, Van Camp G, Huentelman MJ. A sensitive and specific diagnostic test for hearing loss using a microdroplet PCR-based approach and next generation sequencing. Am J Med Genet A. 2013;161A(1):145–152. doi: 10.1002/ajmg.a.35737. [DOI] [PubMed] [Google Scholar]
- 5.Brownstein Z, Friedman LM, Shahin H, Oron-Karni V, Kol N, Abu Rayyan A, Parzefall T, Avraham K. Targeted genomic capture and massively parallel sequencing to identify genes for hereditary hearing loss in Middle Eastern families. Genome Biol. 2011;12(9):R89. doi: 10.1186/gb-2011-12-9-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shearer AE, Hildebrand MS, Smith RJ. Solution-based targeted genomic enrichment for precious DNA samples. BMC Biotechnol. 2012;12:20–6750-12-20. doi: 10.1186/1472-6750-12-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shearer AE, Hildebrand MS, Ravi H, Joshi S, Guiffre AC, Novak B, Happe S, LeProust EM, Smith RJH. Pre-capture multiplexing improves efficiency and cost-effectiveness of targeted genomic enrichment. BMC Genomics. 2012;13(1):618. doi: 10.1186/1471-2164-13-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frommolt P, Abdallah AT, Altmuller J, Motameny S, Thiele H, Becker C, Stemshorn K, Fischer M, Freilinger T, Nürnberg P. Assessing the Enrichment Performance in Targeted Resequencing Experiments. Hum Mutat. 2012;33(4):635–641. doi: 10.1002/humu.22036. [DOI] [PubMed] [Google Scholar]
- 11.Liu X, Jian X, Boerwinkle E. dbNSFP: A lightweight database of human nonsynonymous SNPs and their functional predictions. Hum Mutat. 2011;32(8):894–899. doi: 10.1002/humu.21517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nord AS, Lee M, King M, Walsh T. Accurate and exact CNV identification from targeted high-throughput sequence data. BMC Genomics. 2011;12(1):184. doi: 10.1186/1471-2164-12-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Francey LJ, Conlin LK, Kadesch HE, Clark D, Berrodin D, Sun Y, Glessner J, Hakonarson H, Jalas C, Landau C, Spinner NB, Kenna M, Sagi M, Rehm HL, Krantz ID. Genome-wide SNP genotyping identifies the Stereocilin (STRC) gene as a major contributor to pediatric bilateral sensorineural hearing impairment. Am J Med Genet A. 2011;158(2):298–308. doi: 10.1002/ajmg.a.34391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knijnenburg J, Oberstein SAJL, Frei K, Lucas T, Gijsbers ACJ, Ruivenkamp CAL, Tanke HJ, Szuhai K. A homozygous deletion of a normal variation locus in a patient with hearing loss from non-consanguineous parents. J Med Genet. 2009;46(6):412–417. doi: 10.1136/jmg.2008.063685. [DOI] [PubMed] [Google Scholar]
- 15.Redin C, Le Gras S, Mhamdi O, Geoffroy V, Stoetzel C, Vincent MC, Tanke HJ, Szuhai K. Targeted high-throughput sequencing for diagnosis of genetically heterogeneous diseases: efficient mutation detection in Bardet-Biedl and Alstrom syndromes. J Med Genet. 2012;49(8):502–512. doi: 10.1136/jmedgenet-2012-100875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kohane IS, Hsing M, Kong SW. Taxonomizing, sizing, and overcoming the incidentalome. Genet Med. 2012;14(4):399–404. doi: 10.1038/gim.2011.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tennessen JA, Bigham AW, O’Connor TD, Fu W, Kenny EE, Gravel S, McGee S, Do R, Liu X, Jun G, Kang HM, Jordan D, Leal SM, Gabriel S, Rieder MJ, Abecasis G, Altshuler D, Nickerson DA, Boerwinkle E, Sunyaev S, Bustamante CD, Bamshad MJ, Akey JM. Evolution and Functional Impact of Rare Coding Variation from Deep Sequencing of Human Exomes. Science. 2012;337(6090):64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Korver AMH, Konings S, Dekker FW, Beers M, Wever CC, Frijns JHM, Oudesluys-Murphy AM. Newborn hearing screening vs later hearing screening and developmental outcomes in children with permanent childhood hearing impairment. JAMA: The Journal of the American Medical Association. 2010;304(15):1701–1708. doi: 10.1001/jama.2010.1501. [DOI] [PubMed] [Google Scholar]
- 19.Morton CC, Nance WE. Newborn hearing screening--a silent revolution. N Engl J Med. 2006;354(20):2151–2164. doi: 10.1056/NEJMra050700. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Mapped Sequencing Reads Correlate With 10X Average Depth Of Coverage. 11 outlying samples removed with > 10 Stdev coverage from the mean at 1X, 10X, and 20X.
Figure S2. Variant Discovery is Dependent on a Saturating Depth Of Coverage. Each point represents a single sample data set. Inset shows low coverage data set.
Figure S3. Mapped Sequencing Reads Correlate Linearly With Average Depth of Coverage.
Table S1. General Overview of OtoSCOPE® Platform by Version.
Table S2. Genes Included on OtoSCOPE®.