Abstract
The negatively regulating zinc finger protein (NZFP) is an essential transcription repressor required for early development during gastrulation in Xenopus laevis. In this study, we found that NZFP interacts with the small ubiquitin-like modifier (SUMO) conjugation E2 enzyme, Ubc9, and contains three putative SUMO conjugation sites. Studies with NZFP mutants containing mutations at the putative SUMO conjugation sites showed that these sites were able to be modified independently with SUMO. NZFP was found to be localized in the same nuclear bodies with SUMO-1. However, sumoylation of NZFP did not play a role either in the translocation of NZFP into the nucleus or on nuclear body formation. While wild type NZFP showed significant transcriptional repression, SUMO-conjugation site mutants manifested a decrease in transcriptional repression activity which is reversely proportional to the amount of sumoylation. The sumoylation defective mutant lost its TBP binding activity, while wild type NZFP interacted with TBP and inhibited transcription complex formation. These results strongly suggest that the sumoylation of NZFP facilitates NZFP to bind to TBP and the NZFP/TBP complex then represses the transcription of the target gene by inhibiting basal transcription complex formation.
Keywords: development, NZFP, SUMO, TATA binding protein, Xenopus
INTRODUCTION
During animal development, activation of a series of transcription factors is required. These transcription factors regulate transcription either by activating or repressing their target genes and are maternally expressed or zygotically induced during development (Arce et al., 2006; Buscarlet and Stifani, 2007; Korval, 2007 and references therein). The negatively regulating zinc finger protein (NZFP) is a type of transcriptional repressor that is not only maternally expressed, but is also activated during Xenopus development. After gastrulation, the level of NZFP mRNA decreased significantly between stages 12 and 32. NZFP expression was increased at stage 35 and then began to decrease at stage 48. In adult Xenopus, NZFP is expressed only in the ovary (Kim et al., 2003a; 2003b). NZFP was isolated as a protein that interacts with Xenopus TATA binding protein (TBP) and it is virtually the same protein as XLcGF53.1, which is one of the FAX-ZFP family proteins (Buscarlet and Stifani, 2007; Knöchel et al., 1989). XLcGF53.1 was originally isolated by screening a Xenopus laevis cDNA library specific to t he gastrula stage using the zinc finger sequence as probe (Kim et al., 2003b; Knöchel et al., 1989) and the name, XLcGF53.1, illustrated only the source from which the clone was obtained. We suggested changing the name of XLcGF53.1 to NZFP as the original name is not indicative of its function (Kim et al., 2003b). In addition, although maternally expressed mRNAs of NZFP were maintained until the gastrula stage, NZFP was also induced zygotically at the tadpole stage, i.e., this gene is not gastrula specific (Kim et al., 2003a). NZFP contains a highly conserved sequence designated the finger associated box (FAX) in the N-terminal half and ten C2H2 type zinc finger motifs in the C-terminal half (Kim et al., 2003b). Transcription repression by NZFP is mediated by interaction between F-H boxes of the FAX domain and the C-terminal core domain of TBP which in turn inhibits TFIIA and TFIIB binding to TBP (Kim et al., 2003b).
SUMO-1 is one of four SUMO proteins in mammalian cells and is the most intensively studied member in this class. It is composed of 97-102 amino acids and shares approximately 18% identity with ubiquitin. It can be covalently conjugated to target proteins by a system analogous to the ubiquitin conjugating system (Geiss-Friedlander and Melchoir, 2007; Gill, 2005 and references therein). SUMO-1 is initially activated by Aos1/Uba2 (or SAE1/SAE2) heterodimer (E1 enzyme), which forms a high energy thioester bond with the-SH group of SUMO-1 in an ATP-dependent process. Activated SUMO-1 is transferred to the ubiquitin conjugating E2 enzyme, Ubc9, and then to the amino groups of specific lysine residues of target proteins by forming an isopeptide bond (Gong et al., 1997). Although it was originally proposed that E1 and E2 are enough for sumoylation, some E3-like ligases such as RanBP2, Siz, and PIAS, which are protein inhibitors of activator STATs, were reported to be required for the completion of sumoylation in a similar manner like ubiquitination (Schmidt and Müller, 2003). The consensus sequence of the sumoylation target site is ψKxE, where ψ is the hydrophobic residue, K is the SUMO-1 acceptor lysine, x is any amino acid and E is glutamic acid (Kim et al., 2002; Rodriguez et al., 2001). SUMOs are translated as immature precursors, in which they carry C-terminal extra amino acids (2–11 amino acid residues) that have to be processed by a protease to generate the mature form containing a diglycine motif at its C-terminus (Gareau and Lima, 2010). Proteolytic cleavage of these amino acids is a prerequisite for the conjugation of SUMO to target proteins and is carried out by sentrin-specific protease (SENP). The C-terminal glycine of mature SUMO binds to the amino group of a lysine residue in target proteins. SUMO conjugated target proteins can be desumoylated by SENP that cleaves the bond between glycine within a di-glycine motif of SUMO and the lysine residue of target protein. The free SUMO can be used for another round of sumoylation (Rodriguez et al., 2001).
Sumoylation has been known to play various biological roles including nuclear import of the target protein, control of protein stability, subnuclear localization such as nuclear body formation, and the regulation of transcriptional activity (Kim et al., 2002; Li and Hochstrasser, 2000 and references therein). Sumoylation of transcription factors can control the expression of the target protein largely by repressing transcription (Gill, 2004). Transcription inhibition can be modulated by sumoylation of histone or histone deacetylase (HDAC) interacting proteins. Sumoylation of Elk-1 increased association with HDAC and decreased the acetylation level at the Elk-1 regulated promoter (Yang and Sharrocks, 2004). Histone H4 can also be sumoylated and this sumoylation leads to recruitment of HDAC. Another mechanism to repress transcription is to recruit the corepressor after sumoylation. For example, Daxx binds to the sumoylated Smad4, which has been known to be stimulated by the TGF-β super-family, and represses Smad4-dependent activation (Chang et al., 2005). It was reported that ZNF76, a transcriptional repressor targeting TBP, lost its repression activity by sumoylation (Zheng and Yang, 2004). However, it has not been reported whether sumoylation of a transcriptional repressor induces transcriptional repression activity by facilitating the repressor to interact with TBP.
In this study, we report that NZFP is sumoylated, the sumoylation facilitates NZFP to interact with TBP and the binding of TBP to NZFP results in transcriptional repression.
MATERIALS AND METHODS
Plasmids
For the construction of an expression vector carrying Flag tagged NZFP (NZFP-Flag), the ORF of NZFP was amplified with primers of NZFP-F2 and NZFP-Flag-R containing Flag sequence and the amplified DNA fragments subcloned into the KpnI/BglII sites of pGW1-CMV (British Biotechnology). Site-directed mutagenesis was performed using PCR to generate amino acid substitutions at putative SUMO-1 conjugation sites on NZFP-Flag to construct the NZFP-Flag derivatives; K123R-Flag, K187R-Flag, K233R-Flag, K123RK187R-Flag, K123RK233R-Flag, K187RK233R-Flag and triple mutant-Flag, wherein triple designates the mutant where all three putative sumoylation sites are mutated. The following primers were used for generating mutants: K123R-F, K123R-R, K187R-F, K187R-R, K233R-F, and K233R-R (see Supplementary Table S1 for their sequences).
To fuse His-tag to the N-terminus of NZFP-Flag and its mutants for expression in E. coli, full-length NZFP-Flag and its mutant derivatives were amplified using primers, pRSET-KpnI-NZFP-Flag-F and GW1-R, then inserted into the KpnI/BglII site of pRSET-A vector. pEGFP-C1-NZFP was used (Kim et al., 2003b) to assess subcellular localization. A vector that expresses the GFP-triple fusion protein (pEGFP-C1-triple) was constructed by amplifying DNA containing the triple mutant of NZFP using the same primer used for pEGFP-C1-NZFP and inserting it into BamHI/XbaI sites of pEGFP-C1 (Clontech).
For the transcription assay, a vector (designated pGal4-VP16) that expresses Gal4 DNA binding domain (DBD)-VP16 activating domain (AD) fusion protein was first constructed and then wild type or mutant NZFP was fused to the C-terminus of Gal4-VP16. To generate pGal4-VP16, Gal4 DNA binding domain (DBD) was amplified using primers, Gal4-F and Gal4-R, and inserted into the HindIII/BamHI sites of pcDNA4/TO (Invitrogen) and was designated pGal4-DBD. VP16 activation domain was amplified by using the primers, VP16-F and VP16 (XhoI)-R and inserted into the BamHI/XhoI site of pGal4-DBD. pGal4-VP16-NZFP and derivatives were constructed by insertion of the NZFP and derivatives cDNA into the BamHI/XbaI sites of pGal4-VP16.
To construct HA-TBP wherein the HA tag was added to the N-terminus of TBP, DNA encoding full length TBP was amplified using primers (HA-TBP-F and HA-TBP-R) and inserted into the KpnI and BglII sites of GW1-CMV vector. The sequences of primers used above are shown in Supplementary Table S1. All DNA sequences were determined by Seoul National University Genome Research Facility.
The expression vectors of pcDNA3-myc-SUMO-1, His-Ubc9, GST-SUMO-1, and GST-SAE1/SAE2 were provided by Dr. Chin-Ha Chung (Seoul National University, Korea). The expression vector of pEGFP-SUMO-1 was provided by Dr. Yong-sok Kim (National Institutes of Health, USA). The plasmid for the β-arrestin expression was obtained from Dr. Lefkowitz (Duke University Medical Center, USA).
Antibodies
Rabbit anti-SUMO-1 (Santa Cruz), mouse anti-Flag (Sigma-Aldrich), mouse anti-GFP (Clontech), mouse anti-HA (Sigma-Aldrich), mouse anti-Myc (Santa Cruz) antibodies, anti-mouse Cy3-conjugated IgG, horseradish peroxidase (HRP)-linked goat anti-mouse and anti-rabbit IgG antibodies (Sigma-Aldrich) were purchased commercially.
Cell culture and transfection
293T and COS-7 cells were cultured in DMEM (GIBCO.BRL) supplemented with 10% fetal bovine serum (FBS, Hyclone) and 1% antibiotic-antimycotic liquid (GIBCO.BRL) in 5% CO2 at 37°C. For the co-immunoprecipitation assay to examine SUMO-1 and NZFP interaction, 293T cells were inoculated on 100 mm culture dishes and transfected with 4 μg of pEGFP-SUMO-1 and 8 μg of pGW1-NZFP-Flag and mutants using Lipofectamine-Plus reagent (Invitrogen). For immunostaining assay, COS-7 cells were inoculated on 35 mm culture dishes and transfected with 2 μg of appropriate plasmids using Lipofectamine-Plus reagent. For the transcriptional activity assay, 293T cells were inoculated on 24-well plates (or 15.6 mm culture dishes) and transfected with 400 ng of pGal4-VP16, pGal4-VP16-NZFP or derivatives (effecter plasmids), 200 ng of Myc9-SENP1, Myc9-SENP2 or EGFP-C1 with the combination as indicated in Fig. 5 and 200 ng of pG5TK-luc (reporter plasmid) along with 80 ng of pcDNA4/TO/LacZ (Invitrogen) using the calcium phosphate precipitation method. For the co-immunoprecipitation assay to examine the interaction between TBP and NZFP, 293T cells were inoculated on 6-well plate and transfected with 1 μg of HA-TBP and 8 μg of pGW1-NZFP-Flag and the triple mutant using the calcium phosphate precipitation method.
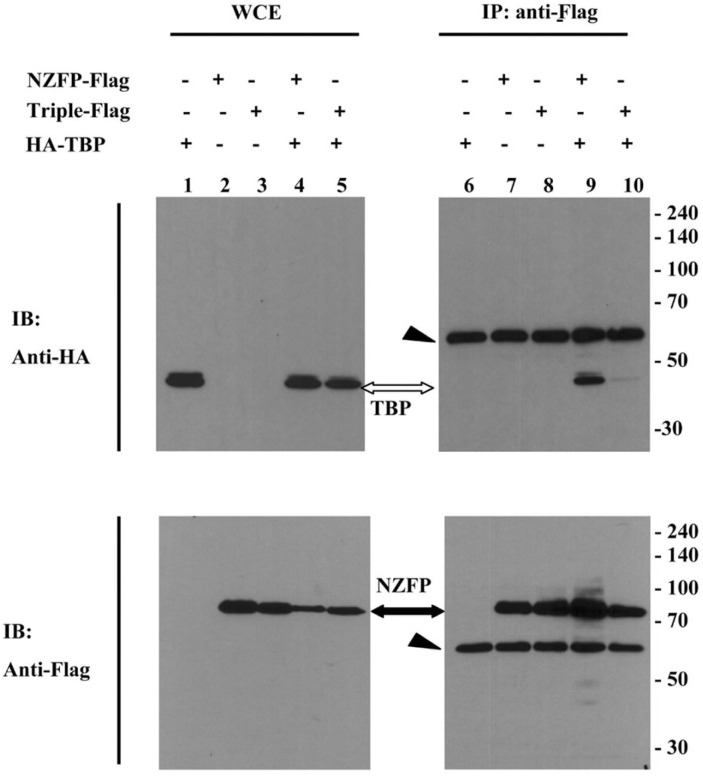
Fig. 5.
Sumoylation of NZFP is required for binding to TBP. 293T cells were transfected with vectors expressing HA-TBP and NZFP-Flag or triple mutant-Flag. Cell extracts were immunoprecipitated using anti-Flag antibody. The whole cell extract (WCE) and bound proteins were analyzed by Western blotting using anti-HA antibody (upper) or anti-Flag antibody (lower) as the primary antibody. The black and white arrows indicate NZFP-Flag and HA-TBP, respectively. The arrowheads represent immunoglobulin heavy chains.
Yeast two-hybrid screen
Yeast two-hybrid screen was carried out in strain L40 as described by the manufacturer’s protocol (Clontech). As bait, the Xenopus NZFP gene was subcloned into the BamHI and PstI sites of the pBAH encoding LexA DNA binding domain. Approximately 107 transformants from a Xenopus oocyte cDNA library (Clontech) were screened with 7 mM of 3-amino-1, 2, 4-triazole (3-AT) containing synthetic medium lacking tryptophan, leucine and histidine for three days. To verify true positive clones, the isolated candidate clones were retransformed into L40 strain with pBAH-NZFP and confirmed by using the β-galactosidase filter assay. The positive clone was subcloned into pBluescript II and sequenced.
In vitro pull-down assay
The 293T cells that can express NZFP-Flag were lysed using RIPA buffer, and cell extracts prepared. GST-Ubc9 fusion protein was expressed in E. coli and purified using glutathione-S-Sepharose beads. The purified GST-Ubc9 was subjected to bind to glutathione Sepharose beads until saturated. NZFP-Flag containing cell extracts and Ubc9 (6 μg) saturated glutathione Sepharose beads were mixed. After constant agitation at 4°C for 4 h, the resin was extensively washed with phosphate buffered saline (PBS). The bound proteins were suspended in 40 μl of SDS-loading buffer and separated on a 10% SDS-PAGE and subject to Western blot analysis.
Transcriptional activity assay
The 293T cells (4 × 105 cells/well) were grown in 24-well dishes for 16 h and transfected with the plasmids as indicated in the legend to Fig. 5. Cell extracts were prepared 24 h after transfection and used in the luciferase assay. The luciferase activities were normalized with β-galactosidase activity. The repression fold was calculated by dividing the normalized luciferase activity of each effector molecule with that of the Gal4-VP16 control. The experiments were performed in triplicate.
Co-immunoprecipitation and Western blot analysis
To check SUMO-1 and NZFP interaction, cell extracts were lysed in SDS lysis buffer (5% SDS, 150 mM Tris-Cl pH 7.5, 30% glycerol), then diluted 1:10 in dilution buffer [0.2% Triton X-100, 20 mM Tris-Cl (pH 7.5), 150 mM NaCl and 1 mM EDTA], containing complete protease inhibitor cocktail (Sigma-Aldrich). After sonication, the lysates were cleared by centrifugation (13, 000 rpm) at 4°C for 30 min. The supernatants were precleared by incubating with protein G agarose (Santa Cruz) for 1 h at 4°C, and then incubated with anti-Flag M2 affinity gel (Sigma-Aldrich) for 3 h at 4°C. The immunocomplexes were washed three times with washing buffer (2% Triton X-100, 20 mM Tris-Cl (pH 7.8), 150 mM NaCl and 1mM EDTA supplemented with complete protease inhibitor cocktail). To check the interaction between TBP and NZFP, cells were harvested, incubated with Co-IP buffer [50 mM Tris-Cl (pH 7.4), 150 mM NaCl, 1 mM EDTA and 1% Triton X-100, 0.1% SDS, 0.8% sodium deoxycholate, 10% glycerol, 1 mM DTT with complete protease inhibitor cocktail] on ice for 15 min and sonicated. The lysates were treated as described above and then incubated with anti-Flag M2 affinity gel overnight at 4°C. The immunocomplexes were washed once with lysis buffer, four times with washing buffer (lysis buffer without SDS), finally washed with washing buffer containing 100 mM of NaCl. The bound proteins were eluted from affinity gel by boiling in gel loading buffer (80 mM Tris-Cl (pH 6.8), 2% sodium dodecyl sulfate, 10% glycerol, 0.01% bromophenol blue, 1% 2-mercaptoethanol). Proteins were subjected to 8% SDS-PAGE and then transferred to a Hybond ECL nitrocellulose membrane (Amersham, GE). Each membrane was treated with the appropriate primary antibodies and then with the HRP-conjugated secondary antibody.
Immunocytochemistry
COS-7 cells were grown on a coverslip and transfected with the myc-SUMO-1 and EGFP-NZFP or EGFP-triple expression vectors. Twenty-four hours after transfection, cells were fixed with a solution containing 4% formaldehyde in PBS for 40 min at room temperature and rinsed with PBS. Fixed cells were incubated with PBS containing 0.1% Triton X-100 for 10 min at room temperature, rinsed with PBS and cells were then blocked with PBS containing 3% BSA (Sigma) at room temperature for 1 h. Cells were then incubated with anti-Myc antibody (1:300 dilution) at room temperature for 1 h. After washing three times with PBS containing 3% BSA, cells were incubated with antimouse Cy3-conjugated secondary antibody (1:1,000 dilution) for 1 h in the dark at room temperature. Confocal laser scanning microscopy was performed with a Leica 2000 microscope, using excitation wavelengths of 543 nm for Cy3 and 488 nm for GFP.
Expression and Purification of Recombinant Proteins
E. coli BL21 (DE3) cells expressing His-Ubc9, GST-SAE1/SAE2 and GST-SUMO-1 were cultured until the OD600 reached 0.8 and expression induced by adding 0.3 mM IPTG (Promega). The expressed proteins were purified using ProBond Purification System (Invitrogen) and glutathione-S-Sepharose beads according to the manufacturer’s instructions (GE Healthcare). The purified proteins were stored in the storage buffer (see Le Drean et al., 2002 for its composition).
To induce the expression of His-NZFP-Flag and its derivatives, cells were incubated overnight at 22°C, after adding 0.3 mM IPTG. The expressed proteins were purified using Ni-NTA agarose and the purified proteins were dialyzed in the protein storage buffer (20 mM HEPES, pH 7.5, 10% glycerol, 5 mM MgCl2, 1 mM DTT).
In vitro sumoylation assay
200 ng of purified target protein, His-NZFP-Flag and its derivates, were incubated with purified 1.5 μg of GST-SAE1/SAE2, 300 ng of His-Ubc9 and 600 ng of GST-SUMO-1 in the 30 μl of reaction system including 2 mM Mg-ATP, 5 mM MgCl2, 20 mM HEPES (pH 7.5), 0.05% Tween-20, 0.1 mg/ml of BSA, 1 mM DTT and protease inhibitors. Reactions were incubated at 37°C for 3 h. After terminating the reaction by adding SDS sample loading buffer containing β-mercaptoethanol, reaction products were fractionated by SDS-PAGE (8%) and then transferred to a Hybond ECL nitrocellulose membrane. Each membrane was treated with the appropriate primary antibodies and then with the HRP-conjugated secondary antibody.
RESULTS
Identification of Ubc9 as a NZFP interacting protein in Xenopus laevis
We have previously shown that NZFP is a novel transcriptional repressor which interacts with TBP and is required for early development during Xenopus gastrulation (Kim et al., 2003b). This previous study suggests that TBP interacting NZFP requires another factor(s) to repress transcription. In order to identify protein(s) that interact(s) with NZFP, yeast two-hybrid screening of a Xenopus oocyte cDNA library was performed. A bait vector was constructed by fusing the gene encoding the DNA binding domain of LexA which is in pBAH to NZFP. The resulting pBAH-NZFP construct was transformed into a yeast strain, L40. Approximately, 1 × 107 transformants from a Xenopus cDNA library were primarily screened by their survival on a 3-aminotriazole containing medium and then by the β-galactosidase assay. Four positive clones were obtained and their sequences determined. Only one of them (clone #3) had a long open reading frame that encoded a protein and clone #3 was confirmed to be a true positive by the β-galactosidase assay (Supplementary Fig. S1A). The sequence of the insert of this clone was identical with that of the Xenopus ubiquitin-like E2 conjugating enzyme, Ubc9. Sequence comparisons of Xenopus Ubc9 (xUbc9) with that of the mammalian homologue showed that they have 100% homology and with Drosophila and yeast homologues 85% and 56% homology, respectively. To confirm that xUbc9 interacts with NZFP, the GST pull-down assay was exploited by using bacterially expressed GST-xUbc9 fusion protein and in vitro translated [35S]-labeled NZFP. As shown in Supplementary Fig. S1B, xUbc9 was bound to NZFP in vitro. The results obtained from the yeast two-hybrid and in vitro GST pull-down assays showed that Ubc9 interacts with NZFP.
Sumoylation of NZFP occurs at all three putative SUMO-1 acceptor sites
As NZFP contains three putative sumoylation sites (see Fig. 1A for their positions) and interacts with Ubc9, it can be assumed that NZFP is possibly modified by SUMO-1. To reveal the sumoylation of NZFP, all the components required for the in vitro sumoylation assay were prepared as fusion proteins from bacteria. As shown in Fig. 1B, incubation of purified His-NZFP-Flag, GST-SUMO-1, His-Ubc9 and GST-SAE1/SAE2 generated retarded bands when either anti-Flag or anti-SUMO antibody was used for Western blot analysis (lanes 6 and 12). These results indicate that there are multiple distinct SUMO-modified NZFPs, suggesting that NZFP contains multiple sumoylation sites (the bracketed areas in Fig. 1B). It should be noted that NZFP is sumoylated only when Ubc9 and SAE1/SAE2 were incubated with SUMO-1 (Fig. 1B, lane 6). Although many retarded bands were generated when NZFP was not incubated, their intensities are weak and their patterns are different from those bands generated by NZFP sumoylation (compare lane 7 with lane 12), suggesting that NZFP is modified by SUMO-1 at multiple sites. To confirm sumoylation of NZFP in vivo, constructs corresponding to NZFP-Flag and GFP-SUMO-1 were cotransfected into 293T cells. After incubating the transfected cells, cell extracts were prepared and Western blot analysis performed (see “Materials and Methods”). When anti-Flag antibody was used as a primary antibody, only the cells cotransfected with EGFP-SUMO-1 and NZFP-Flag showed the slowly migrating bands representing sumoylated NZFP (Fig. 1C, lane 8). However, when anti-GFP antibody was used as a primary antibody, slowly migrating bands were produced in all GFP-SUMO-1-expressing cells, suggesting that EGFP-SUMO-1 is able to modify many other target proteins in the cells (Fig. 1C, lanes 2 and 4). These results indicate that NZFP can be modified by covalent attachment of SUMO-1.
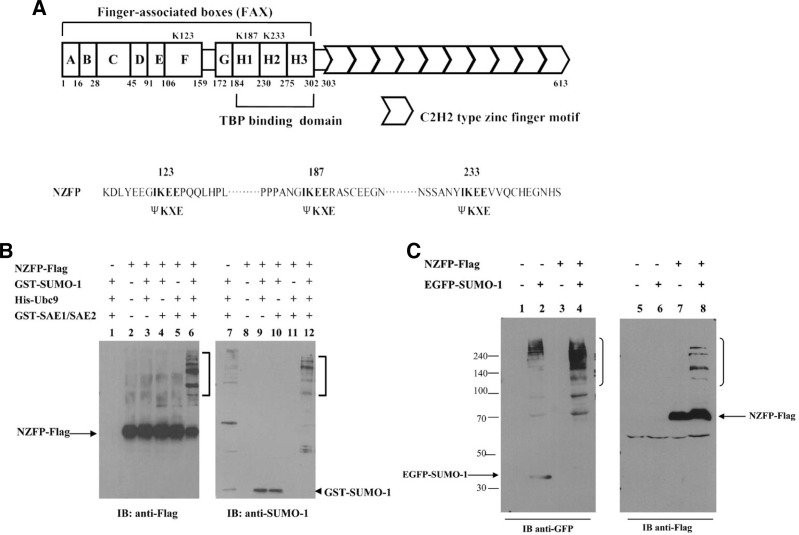
Fig. 1.
NZFP can be modified by SUMO-1 in vitro and in vivo. (A) Schematic representation of NZFP. The positions of putative SUMO-1 acceptor sites (K123, K183, and K233) are marked in FAX region (upper panel). The positions of mutations where the lysine residues within each putative acceptor site were substituted by arginine residues are marked (lower panel). (B) In vitro sumoylation of NZFP. NZFP-Flag, GST-SUMO-1, His-Ubc9, GST-SAE1/SAE2 were expressed in E. coli, purified using affinity chromatography and used in the sumoylation assay. After the reaction was completed, samples were analyzed by Western blotting using antibodies against Flag (left) or SUMO-1 (right). IB designates immunoblotting. Open brackets indicate retarded bands. Unmodified NZFP-Flag and GST-SUMO-1 are indicated by arrow and arrowhead, respectively. (C) In vivo sumoylation of NZFP. 293T cells were transfected with the vectors expressing NZFP-Flag and GFP-SUMO-1, separately or together. Cell lysates were analyzed by Western blotting using antibodies against GFP (left) or Flag (right). The brackets indicate retarded bands produced by sumoylation.
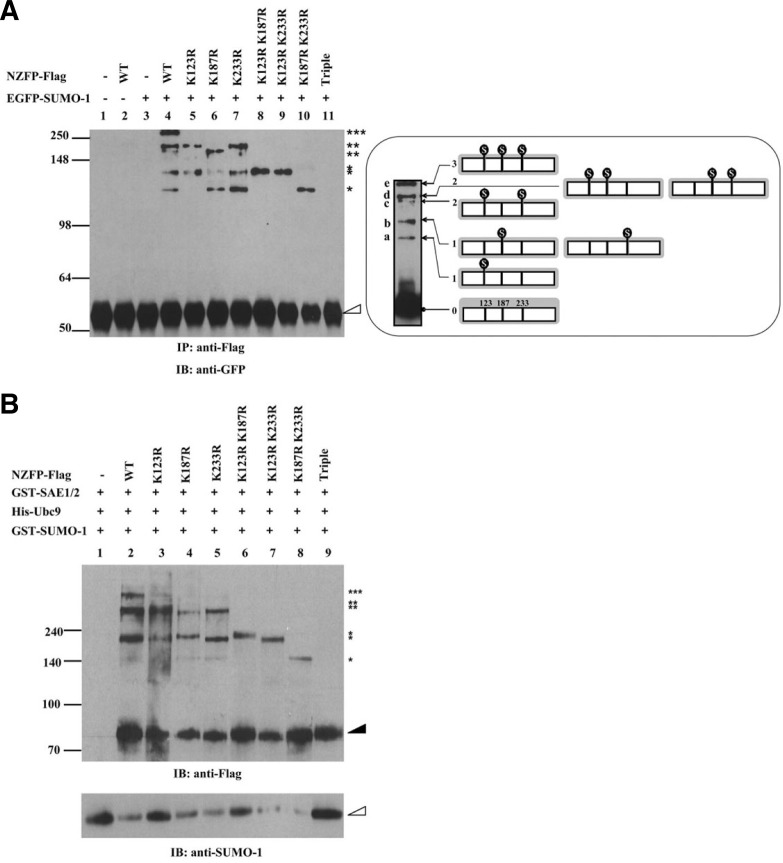
As previously stated, NZFP contains three putative sumoylation sites which correspond to the consensus sequence for sumoylation (Rodriguez et al., 2001). In most cases, sumoylation occurs at the consensus target sequence of ψKxE (Rodriguez et al., 2001). NZFP contains the putative IKEE target sequence at three sites designated as K123, K187 and K233 (Fig. 1A). The sites K123, K187 and K233 are located on the F box, H1 box and H2 box of NZFP, respectively. To determine if SUMO-1 binds to these putative SUMO acceptor sites of NZFP, site-specific mutations were introduced into these sites. Since sumoylation has been reported to occur at the lysine residue in the IKEE consensus sequence, only the lysine was changed to arginine (Lys → Arg) in NZFP. As shown in Fig. 1A, substitutions were performed singularly on one of three putative SUMO-1 acceptor sites (K123R, K187R, or K233R), doubly on two of three putative SUMO-1 acceptor sites (K123RK187R, K123RK233R, or K187RK233R), or simultaneously on the all SUMO-1 acceptor sites (triple mutant). All of mutant types as well as wild type of NZFP were fused to Flag and transfected into 293T cells for immunoprecipitation and Western blot analysis. As shown in Fig. 2A, all three single site mutations (K123R, K187R and K233R) produced three slowly migrating bands, although different combinations of retarded bands appeared. Double mutations (K123RK187R, K123RK233R, and K187RK233R) produced only a single retarded band, indicating that SUMO-1 was bound to only one of three sumoylation sites. However, the triple mutant did not show any retarded band(s), suggesting that all three putative sumoylation sites can be modified by SUMO-1 and there is no more sumoylation site besides these three sites. Based on the analysis of retarded bands corresponding to each mutation, we could identify multiple distinct forms of sumoylated NZFP as schematically represented in the right panel in Fig. 2A. The lowest retarded band (a) represents a single sumoylation of NZFP only at K123. Two other forms (b) of the single sumoylations at K187 or K233 migrated similarly and the uppermost band (e) represents the mature form of sumoylated NZFP at three sites. Figure 2B also supports the above results. Three putative SUMO-1 acceptor sites of NZFP were able to be modified by SUMO-1 in vitro. The band patterns of in vitro experiments are similar to those obtained from in vivo experiments. These studies indicate that NZFP is sumoylated at all putative SUMO-1 acceptor sites (K123, K187 and K233).
Fig. 2.
NZFP is sumoylated at three consensus SUMO acceptor sites. (A) In vivo sumoylation assay of NZFP and its mutant derivatives. Flag fused wild type or mutant NZFP constructs were co-transfected with pGFP-SUMO-1 into 293T cells. Proteins were immunoprecipitated with anti-Flag antibody and the bound proteins were analyzed by Western blotting using anti-GFP antibody (left) as the primary antibody. Asterisks designate the number of SUMO-1 bound to each NZFP. The empty triangle designates immunoglobulin heavy chain. IP and IB designate immunoprecipitation and immunoblotting, respectively. The right panel shows the position of SUMO-1 bound to NZFP (indicated by
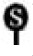 ) and the band produced by each SUMO-1/NZFP complex. Ovoid structure represents SUMO-1 bound to NZFP. (B) In vitro sumoylation of NZFP and its mutants. The proteins described on the top were expressed in E. coli, purified and subjected to the in vitro sumoylation assay. Asterisks represent the number of SUMO-1 bound to each NZFP or its mutants. Solid and empty triangles designate NZFP-Flag and GST-SUMO-1, respectively.
) and the band produced by each SUMO-1/NZFP complex. Ovoid structure represents SUMO-1 bound to NZFP. (B) In vitro sumoylation of NZFP and its mutants. The proteins described on the top were expressed in E. coli, purified and subjected to the in vitro sumoylation assay. Asterisks represent the number of SUMO-1 bound to each NZFP or its mutants. Solid and empty triangles designate NZFP-Flag and GST-SUMO-1, respectively.
NZFP co-localizes with SUMO-1 in nuclear speckle
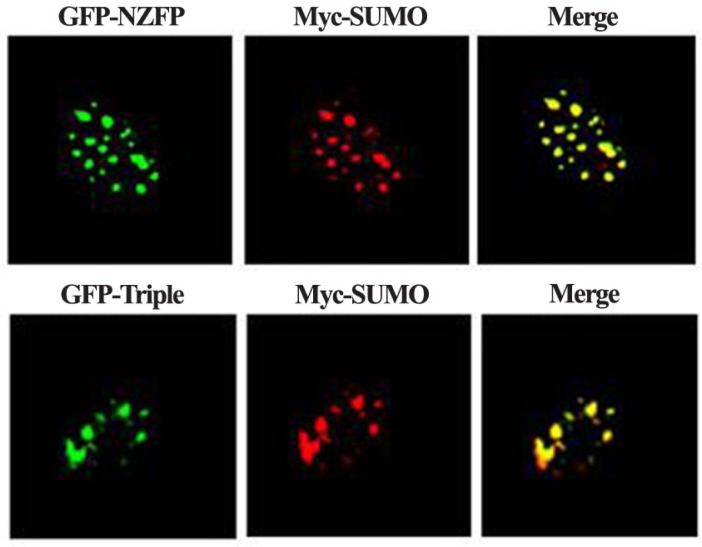
Sumoylation of some target proteins is known to result in the formation of nuclear speckle (Kim et al., 1999; Müller et al., 1998; Zhong et al., 2000). We examined whether NZFP forms nuclear speckles and whether it is localized at the same position with the speckles formed by SUMO-1. Constructs expressing GFP-NZFP and Myc-SUMO-1 were cotransfected into COS-7 cells. After staining the Myc-SUMO-1 with anti-Myc antibody conjugated by Cy3, signals for Cy3 and GFP were detected by confocal laser scanning microscopy. As shown in Fig. 3, wild type NZFP and SUMO-1 were co-localized in nuclear speckles. Surprisingly, the triple mutant was also co-localized with SUMO-1 (lower panel of Fig. 3). These data suggest that, although NZFP and SUMO-1 are co-localized in nuclear speckles, sumoylation of NZFP is not required for speckle formation.
Fig. 3.
Co-localization of SUMO-1 with NZFP in the nuclear speckles. COS-7 cells over-expressing Myc-SUMO-1 and GFP-NZFP or triple mutant of NZFP fused with GFP were fixed and subjected to immunofluorescence staining with anti-Myc monoclonal antibody. The red signal (SUMO-1) was produced by using Cy3-conjugated anti-mouse IgG, whereas the green signal (wild type or mutant form of NZFP) was visualized by GFP fluorescence.
Sumoylation is essential for NZFP to gain its transcriptional repression activity
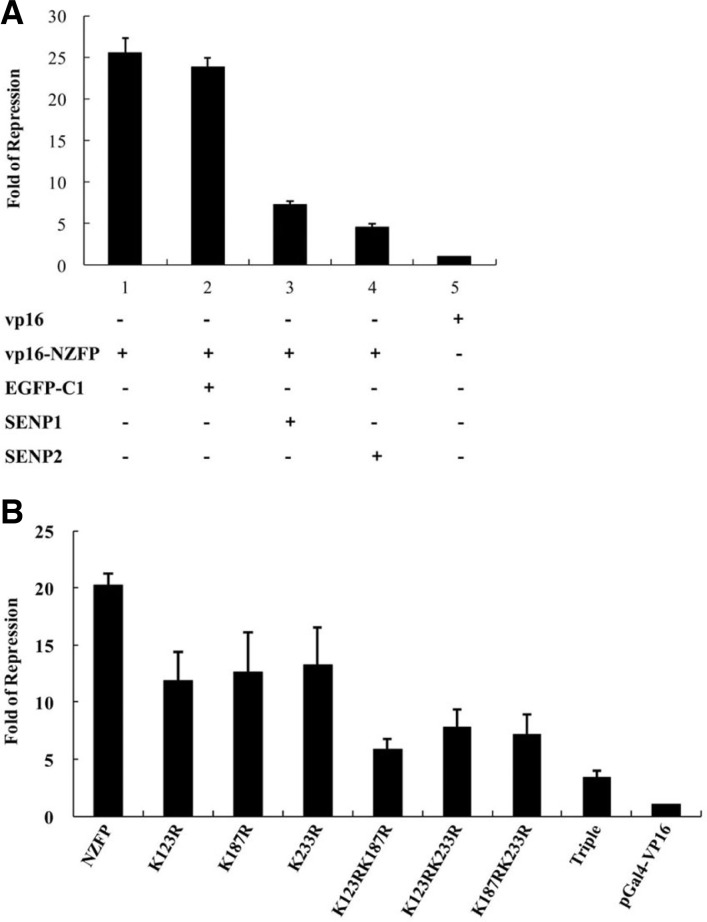
Our previous study showed that the regulatory domain of NZFP is located at its N-terminal half, especially F-and H-boxes which regulate transcriptional activity by interacting with TBP (Kim et al., 2003b). In addition, the sumoylation target sites are also located at the same region. Moreover, the regulation activities of many transcription factors have been shown to be controlled through sumoylation (see “Introduction” and “Discussion”) which raises a question whether sumoylation of NZFP is required for its role in transcriptional repression. To test this possibility, we measured the effect of sumoylation on the transcriptional activity of NZFP. The pG5TK-luc plasmid was used as a reporter to measure transcriptional activity. As the native target genes of NZFP have not been identified, we exploited the Gal4 transcription system as an artificial target gene. Since the basal transcription level of the reporter is very low when only the Gal4 DNA binding domain was used as an effector, the activator domain of VP16 was fused to the C-terminus of Gal4 DNA binding domain to enhance transcriptional activity (designated pGal4-VP16). To express the effector molecules, either wild type or mutant forms of NZFP were fused to the Gal4 DNA binding domain that was also fused to the VP16 activator domain. Both the effector and reporter constructs were co-transfected into HEK 293T cells and the luciferase activities were measured. The repression fold was calculated by dividing the luciferase activity of each experiment with that of VP16. As shown in Fig. 4A, the level of luciferase activity was significantly decreased (repression fold was approximately 25) when wild type NZFP was used as an effector molecule compared to VP16. However, the levels of luciferase activity were significantly recovered by co-transfecting with either SENP1 or SENP2 which is a SUMO-specific protease (only 3–5 fold of repression), while a control backbone vector (EGFP-C1) did not affect transcriptional repression activity. It should be noted that both SENP1 and SENP2 desumoylated NZFP, even though the efficiencies of desumolylation were different (see Supplementary Figs. S2A and S2B). Interestingly, the repression fold was reversely proportional to the efficiency of desumolylation by SENP1 and SENP2 suggesting that transcriptional repression by NZFP is closely related with its sumoylation status (compare Fig. 4A with Supplementary Figs. S2A and S2B). These results strongly suggest that transcriptional repression by NZFP is mediated by the sumoylation of NZFP. To confirm that sumoylation of NZFP is required for the transcriptional repression activity, we examined the effect of sumoylation-site mutations on the luciferase activity. As shown in Fig. 4B, the level of repression was proportional to the degree of sumoylation. Three different forms of single mutant showed 11–13 fold of repression, double mutants 6–8 fold repression, and the triple mutant repressed only 3 folds. It should be noted that the relative luciferase activity produced by the triple mutant is similar to that produced by co-expression of SENP2. These data indicate that NZFP must be sumoylated to repress transcription and sumoylation at all of SUMO-1 acceptor sites is required to obtain the full range of repression activity.
Fig. 4.
SUMO-1 modification is required for transcriptional repression by NZFP. (A) Derepression of gene expression by desumoylation of NZFP. HEK 293T cells were transfected with pcDNA4/TO/LacZ, pG5TK-luc reporter plasmids, and effectors as indicated on the X-axis. The effectors were fused to Gal4DBD-VP16 as described in the text. Cell extracts were prepared at 24 h after transfection and used for luciferase assays. Relative luciferase activities were obtained after normalization with β-galactosidase activity. Fold repression was calculated by dividing the relative luciferase activity of VP16 with that of each effector molecule. Experiments were performed in triplicate and error bars denote the standard deviation from the mean of three independent experiments. EGFP-C1 designates backbone vector used for SENP1 or SENP2 (desumoylating protein) cloning and was used as control. (B) The effect of mutations in SUMO acceptor sites on transcriptional repression activity. Experimental conditions are the same as that used in (A).
Sumoylation facilitates NZFP to interact with TBP
Since transcriptional repression by NZFP is mediated by sumoylation and both the TBP binding site and sumoylation sites are located in the N-terminal region of NZFP, it is intriguing to examine whether sumoylation of NZFP regulates the binding of TBP to NZFP. After constructs corresponding to the Flag-tagged NZFP or triple mutant and the HA-tagged TBP expressing vector were co-transfected into HEK 293T cells, immunoprecipitation using anti-Flag antibody and Western analysis were performed to identify the bound protein. As shown in Fig. 5, the triple mutant in which all three SUMO conjugation sites were mutated lost TBP binding activity, while wild type NZFP binds to TBP (compare lane 9 with lane 10 of upper panel). These studies suggest that sumoylation of NZFP is critical to interact with TBP.
DISCUSSION
Post-translational protein modifications modulate protein function by several means including altering protein activity, altering the ability to interact with ligands, or changing the subcellular localization of the modified protein. Sumoylation is one of the post-translational protein modifications and affects the function of target proteins such as nuclear protein targeting, formation of subnuclear structures, regulation of transcriptional activities or DNA binding abilities of transcription factors, and in addition, control of protein stability (Li and Hochstrasser, 2000). In this study, we discovered that NZFP, a transcriptional repressor which plays key roles during Xenopus early development, was sumoylated at three putative SUMO-1 target sites. Furthermore, transcriptional repression by NZFP is mediated by sumoylation of NZFP and TBP binding to NZFP is facilitated by the sumoylation of NZFP.
In many cases, sumoylation of transcription factors determines the transcriptional regulation status between activation and repression (Fernandez-Martinez et al., 2003). For example, Sp3 is known as an amphifunctional transcription factor, wherein it serves as both activator and repressor. When this protein was sumoylated, it retained only repressor activity (Liu and Shuai, 2008). However, mutation at the SUMO-1 target sites of Sp3 led to the loss of transcriptional repression and to an increase in transcriptional activity (Ross et al., 2002). The androgen receptor (AR) is another example showing the relationship between sumoylation and transcriptional repression. AR is modified by SUMO-1 at its N-terminal domain and sumoylation endows it with transcriptional repression activity (Sapetchnig et al., 2002). In addition, the SUMO-1 acceptor sites in several other transcription factors such as the GR, Myb, and C/EBP have also been mapped to the inhibitory regions of these proteins and mutation of the major SUMO acceptor sites in these factors leads to an increase in transcriptional activation (Bies et al., 2002; Le Drean et al., 2002; Poukka et al., 2000; Tian et al., 2002). However, these reports have not shown that sumoylation of these transcription factors facilitate them to interact with basal transcription factors such as TBP. There are also a few examples where SUMO modification of transcription factors appears to increase transcriptional activity. Post-translational modification of heat shock transcription factors such as HSF1 and HSF2 by SUMO-1 increased the DNA-binding activity of these proteins (Hong et al., 2001; Kim et al., 2002). Another example of transcriptional activation by sumoylation is ZNF76, which is a transcriptional repressor targeting TBP. Sumoylation of ZNF76 loses its activity to bind with TBP and activates transcription (Zheng and Yang, 2004).
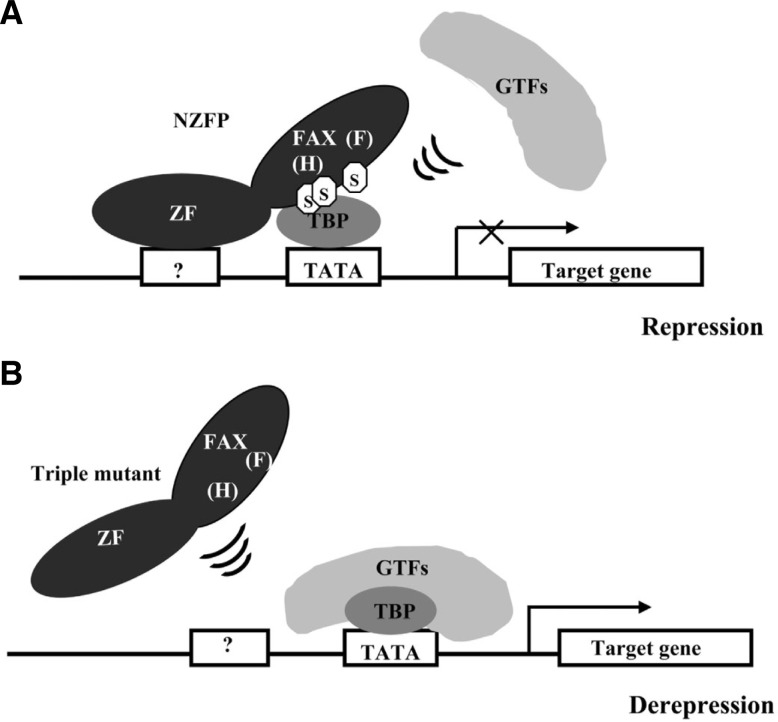
Previously, we reported that the binding of NZFP with TBP inhibits the recruitment of other basal transcription factors such as TFIIA and TFIIB and this binding leads to transcriptional repression (Kim et al., 2003b). In this report, we found that the sumoylation of NZFP enables it to interact with TBP. Considering these results together suggests that there is a novel mechanism of transcriptional repression mediated by sumoylation of a transcription factor: A transcriptional repressor, for example NZFP, is first sumoylated, the sumoylated transcription repressor binds to TBP, and the repressor/TBP complex then inhibits the recruitment of TFIIA and TFIIB at its target gene promoter. However, the unsumoylated form of repressor cannot bind to TBP allowing the general transcription factor complex to be formed at the promoter. Figure 6 illustrates this type of repression using NZFP as an example.
Fig. 6.
Model representing a novel mechanism of transcriptional repression by NZFP. (A) At the transcriptional repression state, NZFP binds to TBP through SUMO modification and the NZFP/SUMO/TBP complex inhibits the binding of other general transcription factors (GTFs) to TBP and leads to transcriptional repression. (B) When NZFP is not modified by SUMO, it is not able to bind to TBP, and thus GTFs bind to TBP and form a transcription initiation complex at the promoter region which mediates the initiation of transcription. Question mark represents an unknown zinc finger binding site. Abbreviations: ZF, zinc finger domain; FAX, FAX domain; H, H-box; F, F-box; S, SUMO.
Supplementary Material
Acknowledgments
We thank Drs. Chin Ha Chung and Yongsok Kim for kindly providing vectors. This work was supported by the Priority Resear-ch Centers Program and Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (Grant Nos 2010-0029694 and 2011-0012947 to BJL) and in part by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research to DLH. MK, ZC, MSS, MSL, JEK, JYK, and JB were supported by Brain Korea 21 Research Fellowship from the Korea Ministry of Education and Human Resources Development.
Note:
Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Arce L., Yokoyama N.N., Waterman M.L. Diversity of LEF/TCF action in development and disease. Oncogene. 2006;25:7492–7504. doi: 10.1038/sj.onc.1210056. [DOI] [PubMed] [Google Scholar]
- Bies J., Markus J., Wolff L. Covalent attachment of the SUMO-1 protein to the negative regulatory domain of the c-Myb transcription factor modifies its stability and transactivation capacity. J. Biol. Chem. 2002;277:8999–9009. doi: 10.1074/jbc.M110453200. [DOI] [PubMed] [Google Scholar]
- Buscarlet M., Stifani S. The ‘Marx’ of Groucho on development and disease. Trends Cell. Biol. 2007;17:353–361. doi: 10.1016/j.tcb.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Chang C.C., Lin D.Y., Fang H.I., Chen R.H., Shih H.M. Daxx mediates the small ubiquitin-like modifier-dependent transcriptional repression of Smad4. J. Biol. Chem. 2005;280:10164–10173. doi: 10.1074/jbc.M409161200. [DOI] [PubMed] [Google Scholar]
- Fernandez-Martinez J., Brown C.V., Diez E., Tilburn J., Arst H.N., Jr., Penalva M.A., Espeso E.A. Overlap of nuclear localisation signal and specific DNA-binding residues within the zinc finger domain of PacC. J. Mol. Biol. 2003;334:667–684. doi: 10.1016/j.jmb.2003.09.072. [DOI] [PubMed] [Google Scholar]
- Gareau J.R., Lima C.D. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell Biol. 2010;11:861–871. doi: 10.1038/nrm3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss-Friedlander R., Melchior F. Concepts in sumoylation: a decade on. Mol. Cell. Biol. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- Gill G. SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes Dev. 2004;18:2046–2059. doi: 10.1101/gad.1214604. [DOI] [PubMed] [Google Scholar]
- Gill G. Something about SUMO inhibits transcription. Curr. Opin. Genet. Dev. 2005;15:536–541. doi: 10.1016/j.gde.2005.07.004. [DOI] [PubMed] [Google Scholar]
- Gong L., Kamitani T., Fujise K., Caskey L.S., Yeh E.T. Preferential interaction of sentrin with a ubiquitin-conjugating enzyme, Ubc9. J. Biol. Chem. 1997;272:28198–28201. doi: 10.1074/jbc.272.45.28198. [DOI] [PubMed] [Google Scholar]
- Hong Y., Rogers R., Matunis M.J., Mayhew C.N., Goodson M.L., Park-Sarge O.K., Sarge K.D., Goodson M. Regulation of heat shock transcription factor 1 by stress-induced SUMO-1 modification. J. Biol. Chem. 2001;276:40263–40267. doi: 10.1074/jbc.M104714200. [DOI] [PubMed] [Google Scholar]
- Kim Y.H., Choi C.Y., Kim Y. Covalent modification of the homeodomain-interacting protein kinase 2 (HIPK2) by the ubiquitin-like protein SUMO-1. Proc. Natl. Acad. Sci USA. 1999;96:12350–12355. doi: 10.1073/pnas.96.22.12350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Cantwell C.A., Johnson P.F., Pfarr C.M., Williams S. C. Transcriptional activity of CCAAT/enhancer-binding proteins is controlled by a conserved inhibitory domain that is a target for sumoylation. J. Biol. Chem. 2002;277:38037–38044. doi: 10.1074/jbc.M207235200. [DOI] [PubMed] [Google Scholar]
- Kim K.I., Baek S.H., Chung C.H. Versatile protein tag, SUMO: Its enzymology and biological function. J. Cell. Physiol. 2002;191:257–268. doi: 10.1002/jcp.10100. [DOI] [PubMed] [Google Scholar]
- Kim M., Choi J., Carlson B.A., Han J.K., Rhee K., Sargent T., Hatfield D.L., Lee B.J. A novel TBP-interacting zinc finger protein functions in early development of Xenopus laevis. Biochem. Biophys. Res. Commun. 2003a;306:1106–1111. doi: 10.1016/s0006-291x(03)01069-6. [DOI] [PubMed] [Google Scholar]
- Kim M., Park C.H., Lee M.S., Carlson B.A., Hatfield D.L., Lee B.J. A novel TBP-interacting zinc finger protein represses transcription by inhibiting the recruitment of TFIIA and TFIIB. Biochem. Biophys. Res. Commun. 2003b;306:231–238. doi: 10.1016/s0006-291x(03)00939-2. [DOI] [PubMed] [Google Scholar]
- Knöchel W., Poting A., Koster M., el Baradi T., Nietfeld W., Bouwmeester T., Pieler T. Evolutionary conserved modules associated with zinc fingers in Xenopus laevis. Proc. Natl. Acad. Sci USA. 1989;86:6097–6100. doi: 10.1073/pnas.86.16.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovall R.A. Structures of CSL, Notch and Mastermind proteins: piecing together an active transcription complex. Curr. Opin. Struct. Biol. 2007;17:117–127. doi: 10.1016/j.sbi.2006.11.004. [DOI] [PubMed] [Google Scholar]
- Le Drean Y., Mincheneau N., Le Goff P., Michel D. Potentiation of glucocorticoid receptor transcriptional activity by sumoylation. Endocrinology. 2002;143:3482–3489. doi: 10.1210/en.2002-220135. [DOI] [PubMed] [Google Scholar]
- Li S.J., Hochstrasser M. The yeast gene ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein. Mol. Cell. Biol. 2000;20:2367–2377. doi: 10.1128/mcb.20.7.2367-2377.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Shuai K. Regulation of the sumoylation system in gene expression. Curr. Opin. Cell Biol. 2008;20:288–293. doi: 10.1016/j.ceb.2008.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller S., Matunis M.J., Dejean A. Conjugation with the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus. EMBO J. 1998;17:61–70. doi: 10.1093/emboj/17.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poukka H., Karvonen U., Janne O.A., Palvimo J.J. Covalent modification of the androgen receptor by small ubiquitin-like modifier 1 (SUMO-1) Proc. Natl. Acad. Sci USA. 2000;97:14145–14150. doi: 10.1073/pnas.97.26.14145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez M.S., Dargemont C., Hay R.T. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J. Biol. Chem. 2001;276:12654–12659. doi: 10.1074/jbc.M009476200. [DOI] [PubMed] [Google Scholar]
- Ross S., Best J.L., Zon L.I., Gill G. SUMO-1 modification represses Sp3 transcriptional activation and modulates its subnuclear localization. Mol Cell. 2002;10:831–842. doi: 10.1016/s1097-2765(02)00682-2. [DOI] [PubMed] [Google Scholar]
- Sapetschnig A., Rischitor G., Braun H., Doll A., Schergaut M., Melchior F., Suske G. Transcription factor Sp3 is silenced through SUMO modification by PIAS1. EMBO J. 2002;21:5206–5215. doi: 10.1093/emboj/cdf510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D., Müller S. PIAS/SUMO: new partners in transcriptional regulation. Mol. Life Sci. 2003;60:2561–2574. doi: 10.1007/s00018-003-3129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian S., Poukka H., Palvimo J.J., Janne O.A. Small ubiquitin-related modifier-1 (SUMO-1) modification of the glucocorticoid receptor. Biochem. J. 2002;367:907–911. doi: 10.1042/BJ20021085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.H., Sharrocks A.D. SUMO promotes HDAC-mediated transcriptional repression. Mol Cell. 2004;13:611–617. doi: 10.1016/s1097-2765(04)00060-7. [DOI] [PubMed] [Google Scholar]
- Zheng G., Yang Y.C. ZNF76, a novel transcriptional repressor targeting TATA-binding protein, is modulated by sumoylation. J. Biol. Chem. 2004;279:42410–42421. doi: 10.1074/jbc.M407287200. [DOI] [PubMed] [Google Scholar]
- Zhong S., Muller S., Ronchetti S., Freemont P.S., Dejean A., Pandolfi P.P. Role of SUMO-1-modified PML in nuclear body formation. Blood. 2000;95:2748–2752. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.