Figure 1.
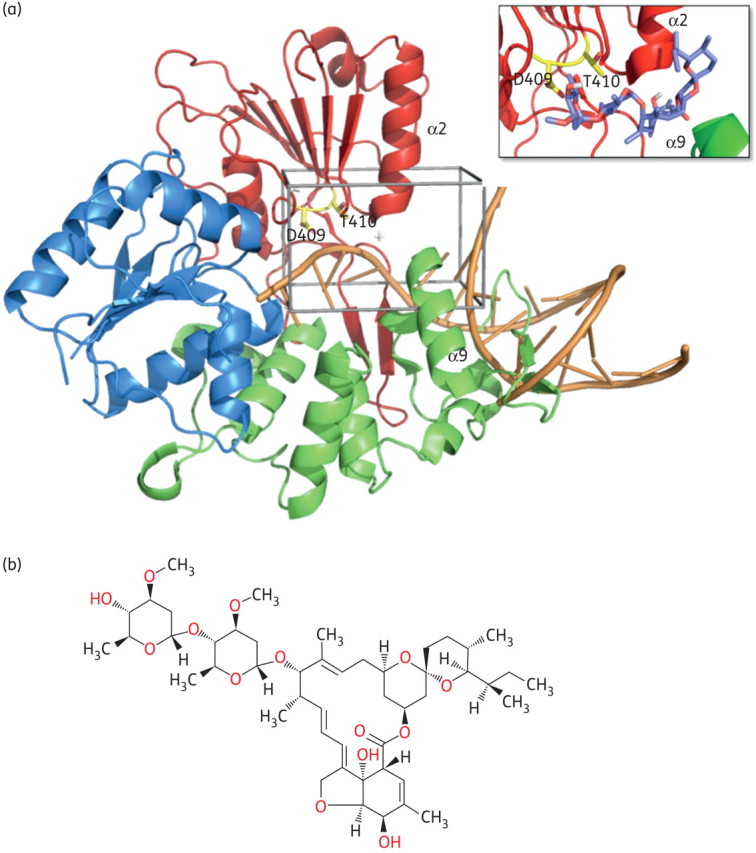
In silico docking of flavivirus helicase. (a) Model of the helicase/RNA interaction for WNV helicase domain. The crystal structure of free WNV helicase (PDB 2QEQ) was used to identify a potential ssRNA access site to the enzyme active site by superposition with the DNA-bound bacterial helicase PcrA (PDB 3PJR). The respective region is located between helices α2 in subdomain II (red) and α9 in subdomain III (green) of the WNV helicase, forming an α-helical gate for the entering RNA substrate (orange worm/sticks). This putative helicase ssRNA access site (enclosed in black) was then chosen as the target site for the in silico ligand search. The amino acids selected for mutation (Asp409 and Thr410) are shown in yellow. The close-up box shows a potential docking conformation of ivermectin in the ssRNA access site. The location of the molecule's macro ring is well established, having the two sugar rings of the compound located inside the protein. The region occupied by the inhibitor is conserved in all binding modes of the different docked conformations. (b) Chemical structure of ivermectin (B1a form; Sigma-Aldrich). Figures created using PyMol (http://www.pymol.org). This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
