Figure 1.
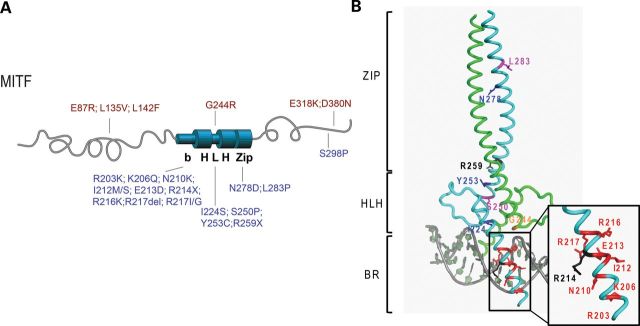
MITF mutations found in melanoma, WS2A and TS patients. (A) A graphic depiction of the human MITF protein with the location of the mutations indicated. The mutations found in melanoma patients are indicated in red (on top). These mutations are located at the amino- and carboxyl ends, and only one mutation is located in the helix–loop–helix domain (G244R). The mutations found in WS2A and TS patients are indicated in blue (on the bottom). Most of these mutations are located in the BR of MITF and in the HLH and zipper domains. The WS2A mutation S298P is located at the carboxyl end of MITF. (B) Structure of the bHLHZip region of MITF showing the location of the mutant residues. To separate mutations leading to TS and WS2A syndromes and those causing melanoma, the respective mutations are mapped on two different molecules of the dimeric MITF structure. The TS/WS2A mutations on the first protomer in cyan are highlighted in the following colors: residues mutated into stop codons, black; residues mutated into prolines, magenta; residues when mutated affecting direct DNA contacts, red; residues when mutated affecting protein–protein interactions, blue. In sharp contrast, only one melanoma-inducing mutation has been found within the bHLHZip region for which the structure of MITF was determined. This mutation (G244) has been highlighted in orange on the second MITF protomer, colored in green.
