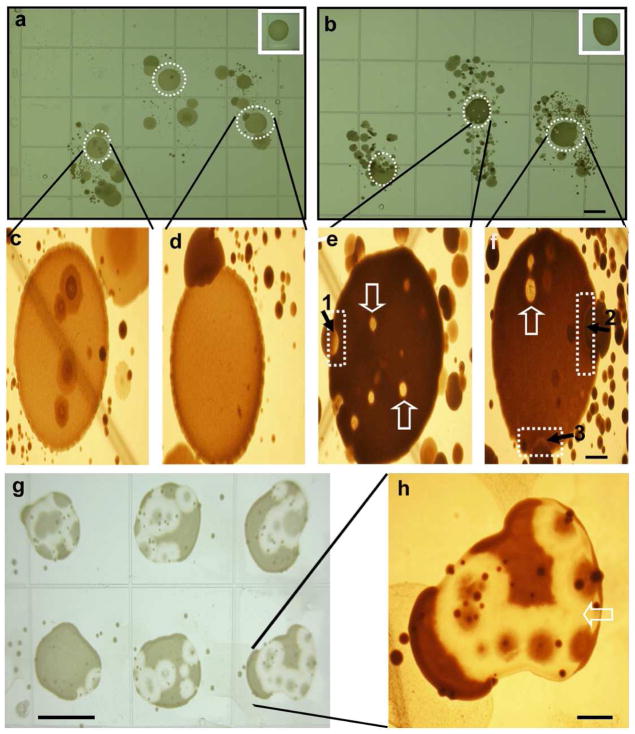
Figure 2. Skin fingerprint analysis of glycerol fermentation of skin microorganisms against P. acnes.
The fingerprints of index, middle, and ring fingers were pressed onto the surface of rich medium agar plates in the absence (a) or presence (b) of 20 g/l glycerol at 30°C unde r anaerobic conditions using Gas-Paks. P. acnes (107 CFU in 5 μl PBS) was spotted on the central portion of each fingerprint. Spotting P. acnes away from fingerprints served as controls (inserts). The high magnitude photos of (a) and (b) were displayed in (c, d) and (e, f), respectively. The inhibition zones (dash squares in e and f) were detected at the boundary between colonies of P. acnes and skin microorganisms. The bubble-like territories of competition (open arrows) were found within P. acnes colonies. In a representative plate, single colonies labeled 1 and 2 (solid arrows) were identified as S. epidermidis. A single colony labeled 3 was identified as Paenibacillus sp. Y412MC1. Six additional colonies from fingerprint bacteria of two different subjects were identified as S. epidermidis. S. epidermidis (105 CFU in 100μl PBS) from colony 1 was re-streaked on an agar plate containing glycerol followed by spotting six separate drops of P. acnes (107 CFU in 5 μl PBS) on the top of a S. epidermidis streak (g). A high magnitude photo of one of P. acnes colonies (g) was displayed in (h). Bars (a-b, g)=0.5 cm; (c-f, h)=0.1 cm.