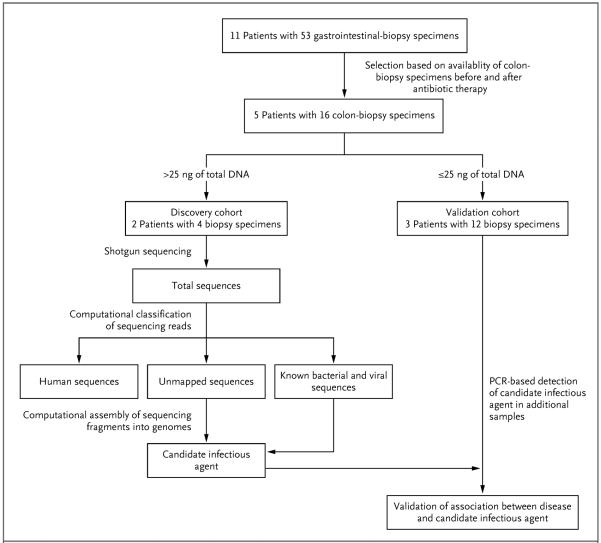
Figure 1. Sample Selection and Analyses.
Formalin-fixed, paraffin-embedded samples obtained from patients with cord colitis were selected for molecular analysis on the basis of clinical criteria. Patients for whom colon-biopsy samples were available for the period ranging from 120 days before antibiotic therapy to 200 days after such therapy were selected for inclusion in the initial cohort. DNA extraction and sequencing were followed by PathSeq analysis,19 in which computational subtraction was applied for the removal of human and known microbial sequences. The remaining unmapped sequencing reads and the reads with a high degree of homology with known microbial sequences were then computationally assembled into longer contiguous overlapping sequences (contigs) representing genomic fragments of a novel organism. Candidate pathogens, which were predicted by PathSeq analysis of the discovery cohort, were detected by targeted methods, such as polymerase-chain-reaction (PCR) assay, in the validation cohort.