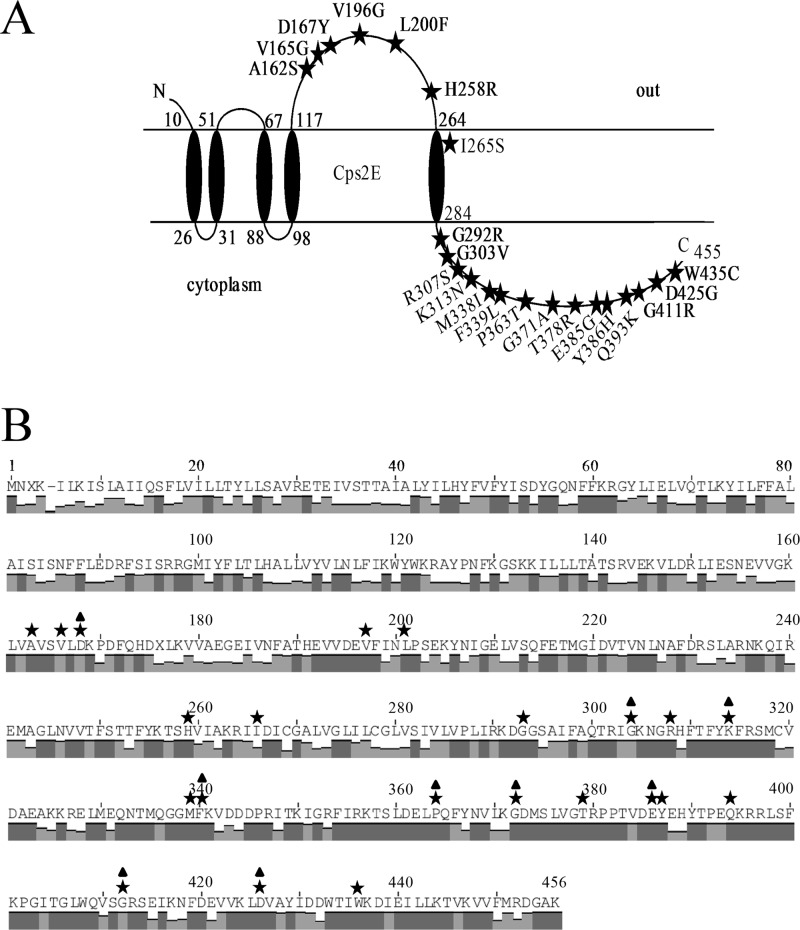
Fig 5.
S. pneumoniae Cps2E topology and CpsE consensus sequence. (A) S. pneumoniae Cps2E topology and suppressor mutations (25, 34). The topology was determined using TMpred program from the ExPASy Proteomics website (http://www.expasy.org/tools), as previously reported (34). Only missense mutations are shown. The P363T and T378R mutations were each identified in two independent mutants. Five nonsense mutations (E191, L244, L369, W407, and Q408) and three frameshift mutations (M59, E240, and G293) have also been identified. In the original publication (34), L200F and K313N were incorrectly listed as L199F and K312N, respectively. (B) Conservation of Cps2E homologues in S. pneumoniae and other bacteria. Amino acid sequences from the 69 S. pneumoniae serotypes containing Cps2E homologues were used in a ClustalW alignment to generate a consensus sequence. Bars indicate the degree of conservation for the respective residues, where the height of the bar correlates to a greater conservation (0 to 100%; dark gray bars indicate 100% conservation). Stars above amino acids indicate relative positions of residues in the consensus CpsE where suppressor mutations were identified in the S. pneumoniae Cps2E. Triangles indicate residues conserved among Cps2E homologues in multiple other bacteria, including Clostridium phytofermentans ISDg (GenBank accession number YP_001558318), Clostridium bolteae (WP_002576468), Streptococcus mitis (WP_001013652), Streptococcus oralis (BAD22619), and Streptococcus parasanguinis (WP_003009077). Twelve residues (A177, D182, G310, G321, K331, F361, P385, G393, E407, G433, D446, and W467) are conserved in WbaP from Salmonella enterica subsp. enterica serovar 4 (YP_008266333). Note that residue numbers are shifted by one from panel A due to insertion of a gap (denoted by a hyphen) for alignment of consensus sequence. X, no conservation of residue.