Fig 3.
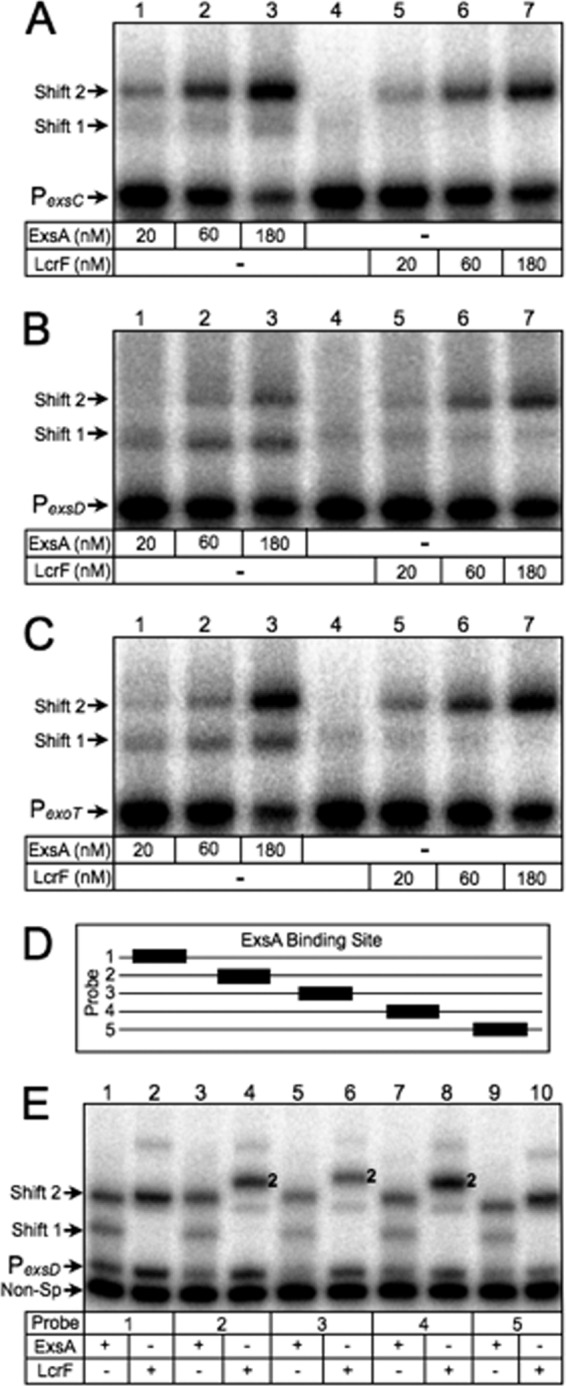
DNA bending properties of ExsA and LcrF. (A to C) EMSAs using 50-bp radiolabeled probes derived from the ExsA-dependent PexsC (A), PexsD (B), and PexoT (C) promoters. Probes (0.05 nM each) were incubated in the presence of 20, 60, or 180 nM ExsA (lanes 1 to 3 in each panel) or LcrF (lanes 5 to 7 in each panel) for 15 min at 25°C. Samples were analyzed by native polyacrylamide gel electrophoresis and phosphorimaging. (D) Diagram depicting the position of the ExsA binding site (black box) derived from the PexsD promoter within probes 1 to 5 (solid line). (E) Circular permutation experiment performed using probes 1 to 5 (0.05 nM each) incubated in the presence of 180 nM ExsA (odd-numbered lanes) or LcrF (even-numbered lanes) for 15 min at 25°C. Samples were analyzed by native polyacrylamide gel electrophoresis and phosphorimaging. The positions of shift products 1 and 2 are indicated.
