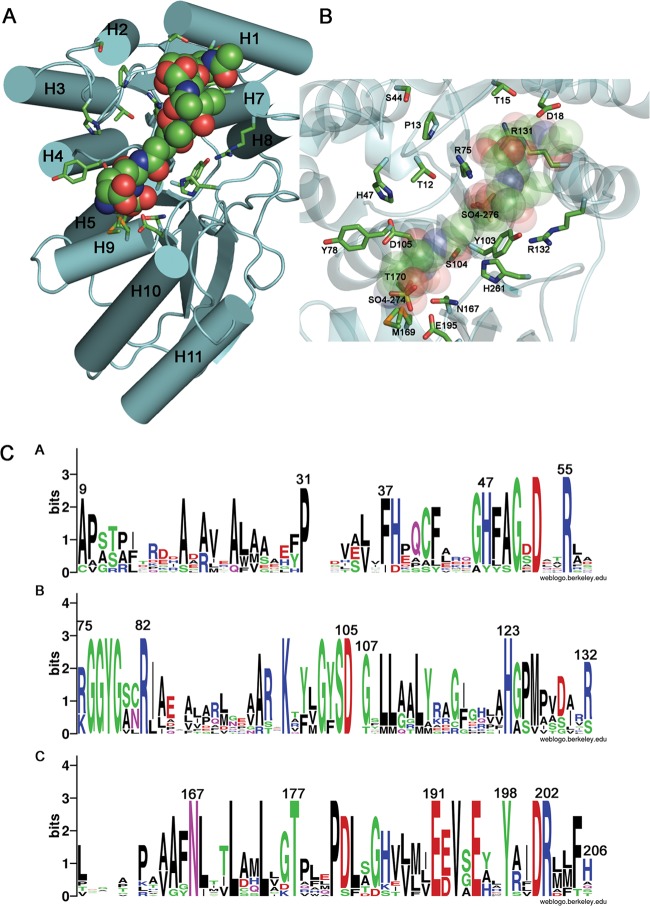
Fig 4.
Model of protein-ligand complex and conservation of putative functional residues. (A) The binding of NaLdcA to the ligand GlcNAc-MurNAc-l-Ala-γ-d-Glu-meso-A2pm-d-Ala (PDB ligand ID MLD, from PDB ID 2cb3) was modeled such that the carboxy-terminal d-Ala is positioned for catalysis near the active site Ser-His-Glu triad. The molecular orientation is similar to that in Fig. 3, and the helices are labeled. (B) Magnified view of the binding site. Oxygen atoms from the carboxylate groups of A2pm and d-Glu of the ligand are superimposed on the two sulfate ions (SO4-274 and SO4-276, respectively) present in the substrate-binding pocket and present similar interactions to the active site residues Arg75 and Glu195, thereby suggesting that the sulfate ions mimic portions of the substrate. (C) Sequence logo representation of relative frequencies of occurrence of residues in NaLdcA and other homologs, obtained using Weblogo (68; http://weblogo.berkeley.edu/). The same set of proteins and multiple-sequence alignment listed in Fig. 3 were used in this analysis. Panels A, B, and C represent different sequence ranges in the enzyme.