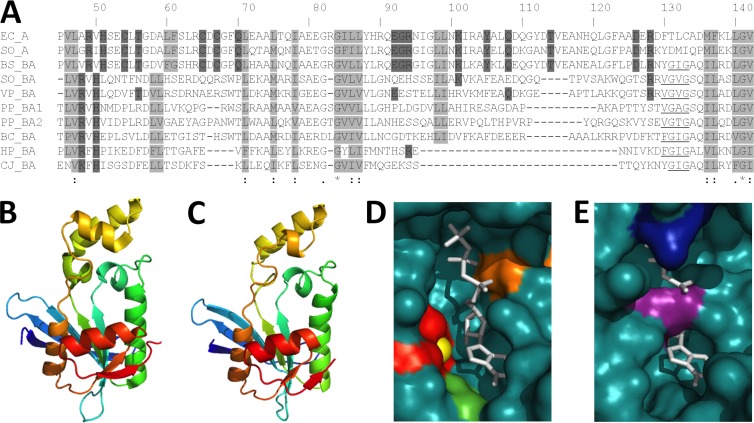
Fig 3.
Alignment and structural modeling of GTP cyclohydrolase II domains from Proteobacteria. (A) ClustalW alignment of the E. coli and MR-1 RibA sequences and the RibBA-CTD sequences of select Proteobacteria. Based on the published crystal structure of E. coli RibA (28), nonpolar residues that form the hydrophobic core are boxed in light gray, and residues that directly participate in the catalytic mechanism are boxed in dark gray. The underlined stretch of nonpolar amino acids is conserved only in the RibBA sequences. Numbering is based on the E. coli RibA sequence. (B to E) Tertiary structure prediction of the RibBA-CTD from MR-1 (C), based upon the crystal structure of RibA from E. coli (B). The RibBA-CTD (residues 188 to 367) was threaded onto the crystal structure of RibA by using PHYRE2 (www.sbg.bio.ic.ac.uk/phyre2/), and the resulting model was visualized using PyMOL (www.pymol.org). The active sites of RibA (D) and the RibBA-CTD (E) are displayed using space-filling models. The GTP analog phosphomethylphosphonic acid guanylate ester (GMPCPP), cocrystallized with RibA from E. coli, is shown in white and superimposed onto the RibBA-CTD for comparison. Active site residues Cys54, Cys65, and Cys67 (red), a coordinated zinc ion (yellow), Tyr105 (green), and Arg128 (orange) are also indicated. The active site residues and coordinated zinc ion are not highlighted in panel E because they are not present in the RibBA-CTD. Arg269 (purple) and Arg350 (blue), the residues extending into the binding pocket of the RibBA-CTD, are indicated instead.