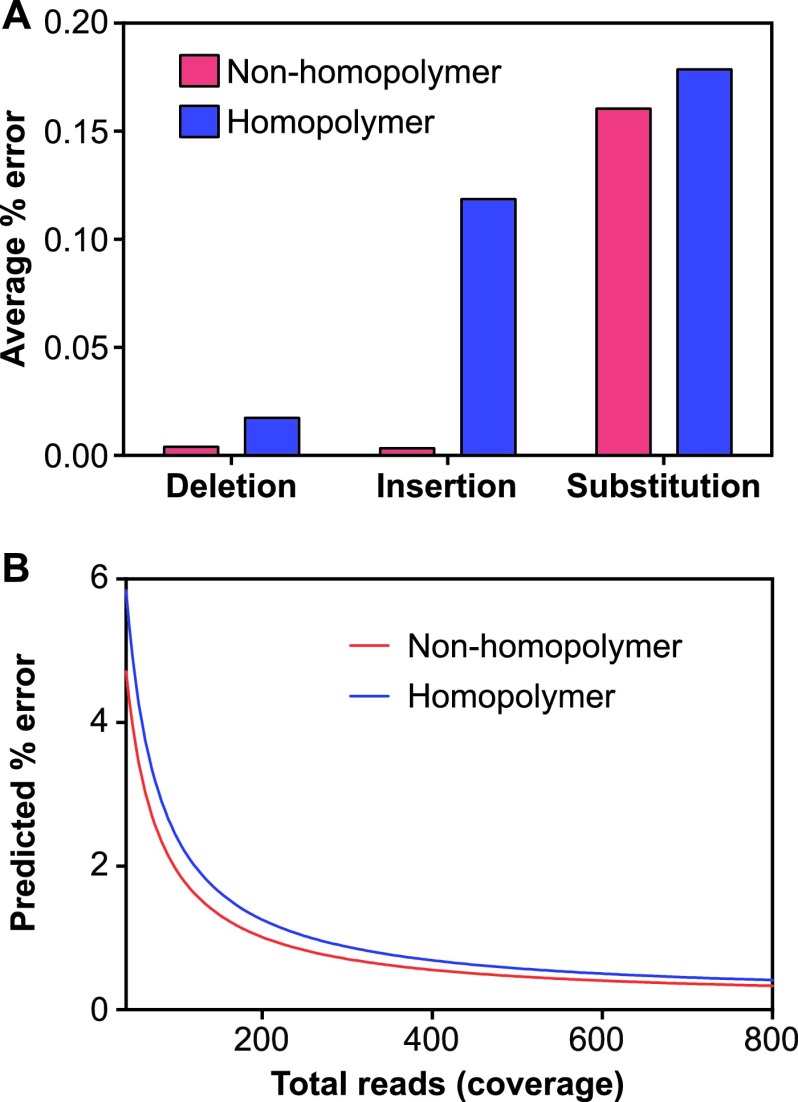
Fig 3.
Error rates for plasmid clones in homopolymer and nonhomopolymer regions. (A) Observed error rates per error type and sequence context. UL54 and UL97 plasmid clones mixed at 1,000 copies/ml plasma were processed and sequenced in duplicate. All variant reads called by the GS amplicon variant analyzer compared to Sanger sequencing of the plasmids were considered errors and were calculated as the percentage of the total reads covering a given nucleotide. The data are presented as the average percent error for substitutions, insertions, and deletions in the homopolymer and nonhomopolymer regions. The averages from two independent experiments are shown. (B) Threshold for minor variant detection relative to total read coverage based on empirical error rates for plasmid clones. The covariates included in the negative binomial model were total_reads (number of reads covering a given nucleotide position), copy number (1,000 or 10,000 copies/ml plasma), and location in homopolymer versus nonhomopolymer regions. The average from two independent experiments is shown.