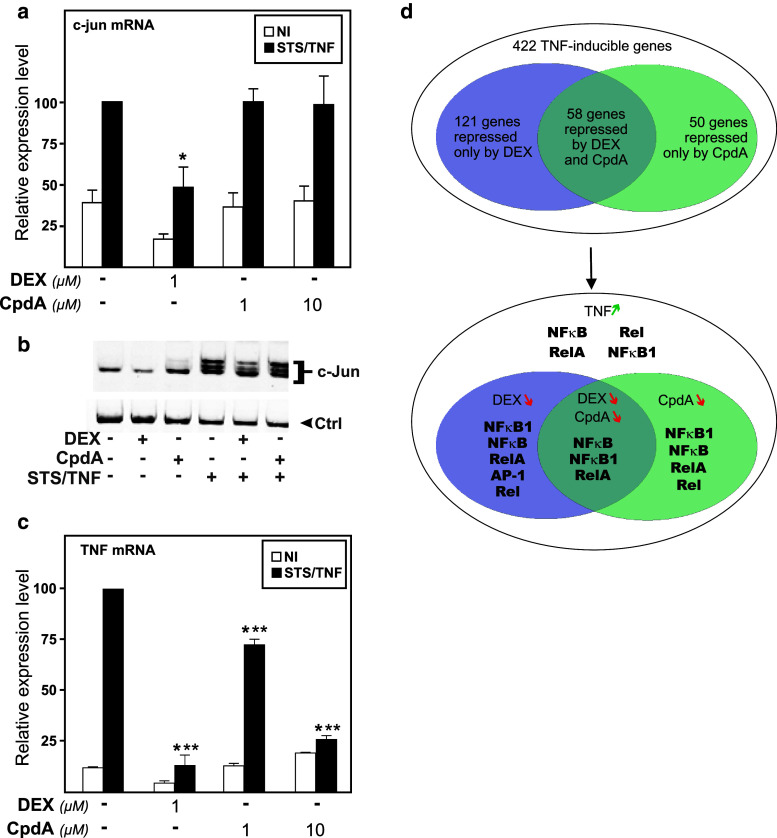
Fig. 4.
Predominantly AP-1-regulated target genes are only transrepressed by DEX- and not by CpdA-activated GR, but promoter complexity determines the final outcome. a, c L929sA cells, starved for 48 h in DMEM devoid of serum, were pre-incubated with solvent, DEX (1 μM), or CpdA (1 or 10 μM) for 1 h, before STS (60 nM) and TNF (2,000 IU/ml) were added, where indicated, for 6 h. Total RNA was isolated and subjected to RT-qPCR assaying cellular c-jun, TNFα, and β-actin and hypoxanthine–guanine phosphoribosyltransferase (HPRT) household gene mRNA levels. Specific signal for cDNA of c-jun or TNFα was normalized to the averaged household genes signal. The STS/TNF condition was set at 100 and all other conditions were recalculated accordingly to allow ratio comparisons. Total solvent concentration was kept similar in all conditions. Results are shown ± SD. The experiment was carried out at least in triplicate and the results are averages of at least two independent experiments. Results of the statistical analysis via ANOVA followed by a Tukey multiple comparison post-test are shown for particular groups of interest, in comparison to the STS/TNF group). b L929sA cells, starved for 48 h in DMEM devoid of serum, were treated with solvent, a combination of STS (60 nM) and TNF (2,000 IU/ml) for 5 h in absence or presence of a 1 h pretreatment of DEX (1 μM) or CpdA (10 μM). Total protein extracts were prepared in duplicate and subjected to Western-blot analysis to detect c-Jun protein. Detection of NF-κB p65 served as a loading control (Ctrl). The result is a representative of three independently performed experiments. d A549 cells, starved for 24 h in DMEM devoid of serum, were pretreated for 1 h either with solvent, DEX (1 μM) or CpdA (10 μM), either or not followed by a 3-h treatment with TNF (2,000 IU/ml). Gene expression levels of corresponding RNA samples were evaluated by a whole genome transcriptome Agilent array (upper panel). Genes with adjusted p values lower than 0.05 in at least one contrast and a fold change higher than 1.3 were selected as significant. Pscan analysis with a minimal statistical significance of p < 0.05, indicates enrichment for specific transcription factor binding motifs in the corresponding gene promoters. Here, we mention the identified NF-κB and AP-1 family members in bold (lower panel)