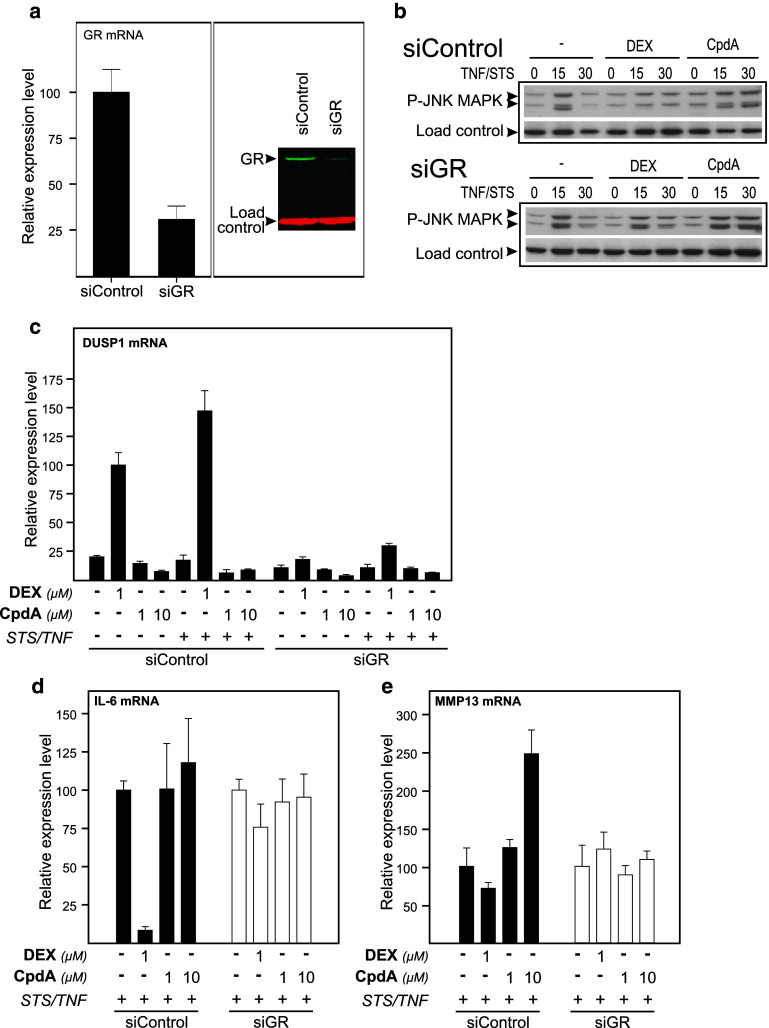
Fig. 8.
GR is essential to mediate the gene expression modulation effect of DEX and CpdA. L929sA cells were transfected with siRNA control (siControl) or siRNA targeted at GR (siGR) and were allowed to rest for 48 h post transfection. In the 16-h period before induction or sampling, cells were starved in DMEM devoid of serum. a We controlled for the efficiency of siRNA GR targeting. Total RNA was isolated and subjected to RT-qPCR for GR mRNA levels and expression levels were normalized to housekeeping gene controls. The expression levels for GR (siControl) were set at 100 and the siGR condition was recalculated accordingly (left panel). Cell lysates were made and GR protein was visualized via Western-blot analysis (right panel). Actin served as a loading control. b SiRNA-transfected L929sA cells (siControl or siGR) were pretreated with solvent, 1 μM DEX, or 10 μM CpdA for 1 h, followed by either or not TNF (2,000 IU/ml) combined with STS (60 nM), for the indicated time points (in minutes). Cell lysates were made and activated JNK was detected using the phospho-specific JNK MAPK antibody. NF-κB p65 served as a loading control (indicated as load control). c–e SiRNA-transfected L929sA cells (siControl or siGR) were pre-incubated with solvent, DEX (1 μM), or CpdA (1 or 10 μM) for 1 h, before TNF (2,000 IU/ml) and STS (60 nM) was added, where indicated, for 6 h. Total RNA was isolated and subjected to RT-qPCR for specific target genes, and expression levels in each treatment group were normalized to housekeeping gene controls. Normalized mRNA levels for DUSP1 (c) was presented with the DEX (SiControl) set as 100 and all other conditions were recalculated accordingly. Normalized mRNA levels for IL-6 (d) and MMP13 (e) were presented with the STS/TNF condition (siControl and siGR) set at 100 and all other conditions were recalculated accordingly to allow ratio comparisons. Total solvent concentration was kept similar in all conditions. Results are shown ± SD. The qPCR was carried out at least in triplicate. Results are representative for two independent experiments