Abstract
Decaprenylphosphoryl-d-arabinofuranosyl (DPA), the immediate donor for the polymerized d-Araf residues of mycobacterial arabinan, is synthesized from 5-phosphoribose-1-diphosphate (PRPP) in three-step reactions. (i) PRPP is transferred to decaprenyl-phosphate (DP) to form decaprenylphosphoryl-d-5-phosphoribose (DPPR). (ii) DPPR is dephosphorylated to form decaprenylphosphoryl-d-ribose (DPR). (iii) DPR is formed to DPA by the epimerase. Mycobacterium tuberculosis Rv3806c and heteromeric Rv3790/Rv3791 have been identified as the PRPP: decaprenyl-phosphate 5-phosphoribosyltransferase and the epimerase respectively. Rv3807c, however, as the candidate of phospholipid phosphatase, catalyzing the biosynthesis of decapreny-l-phosphoryl-ribose (DPR) from decaprenylphosphoryl-β-d-5-phosphoribose by dephosphorylating, has no direct experimental evidence of its essentiality in any species of mycobacterium. In this study, Rv3807c gene was amplified from the genome of M. tuberculosis H37Rv by PCR, and was successfully expressed in Escherichia coli BL21 (DE3) via the recombinant plasmid pColdII-Rv3807c. The resulting protein with the 6× His-tag was identified by SDS-PAGE and Western blotting. The protein was predicted through bioinformatics to contain three transmembrane domains, the N-terminal peptide, and a core structure with phosphatidic acid phosphatase type2/haloperoxidase. This study provides biochemical and bioinformatics evidence for the importance of Rv3807c in mycobacteria, and further functional studies will be conducted for validating Rv3807c as a promising phospholipid phosphatase in the synthetic pathway of DPA.
Keywords: Mycobacterium tuberculosis, Rv3807c, Decaprenylphosphoryl-d-arabinose (DPA)
Introduction
The cell wall core of Mycobacterium tuberculosis is termed the mycolyl-arabinagalactan-peptidoglycan (mAGP) complex, which is essential for the viability of M. tuberculosis. Beyond the membrane is peptidoglycan (PG) in covalent attachment to arabinogalactan (AG), which in turn is attached to the mycolic acids with their long meromycolate and short α-chains [1]. The AG is the major cell wall polysaccharide of mycobacteria. This unique nature leads to the prediction that the enzymes that synthesize this structure should yield a number of potential drug targets [2, 3].
The d-arabinofuran residues (Araf) and d-galactofuran residues (Galf) are added to form the mature lipid-linked AG, and the immediate donor of the polymerized Araf is decaprenylphosphoryl-d-arabinose (DPA) contributing to AG and lipoarabinomannan (LAM) assembly [3–5]. Many of the enzymes catalyzing the synthesis of DPA serving as an arabinose donor in the cell wall biosynthesis have been identified [6] and utilized as the targets for the development of new drugs against TB [7, 8].
Decaprenylphosphoryl-d-arabinose is synthesized initially from the transfer of phosphoribosyl pyrophosphate (PRPP) to decaprenyl phosphate (DP) to form 5-phospho-decaprenylphospho-ribose (5-p-DPR) via the 5-phosphorobosyl transferase (Rv3806c) [9]. Subsequently, the 5-phosphate unit is removed from 5-phospho-decaprenylphospho-ribose to form decaprenyl-phosphoryl-ribose (DPR) by the putative Rv3807 phospholipid phosphatase [10, 11]. The final step of the biosynthetic pathway of decaprenyl-phospho-arabinose is the 2′-epimerization from DPR to DPA, catalyzed by the heteromeric Rv3970/Rv3971 2′-epimerase which is probably not released from the mycobacterial enzyme under physiological conditions [12, 13]. The function and characteristics of the 5-phospho-α-d-ribose-1-pyrophosphate: decapronyl phosphate 5-phosphoribosyltransferase (Rv3806c) and the downstream enzyme decaprenyl-phospho-ribose-2′-epimerase (Rv3790/Rv3791) are identified to be essential for synthesize of decapresyl-phospho-arabinose (DPA). However, the biological function of the Rv3807 gene product, as a good candidate of a specific decaprenyl-phospho-ribose-5′-phosphate phosphatase, which is located next to the phosphoribosyltransferase (Rv3806c), has not been accurately elucidated yet. The upstream Rv3806c is reversible in the absence of pyrophosphatase activity, therefore the phosphatase reaction is expected to be irreversible [11]. It is necessary to identify the phospholipid phosphatase (Rv3807c) and to accomplish the mechanism of biosynthesis of decaprenyl-phospho-arabinose in mycobacteria.
The Rv3807c orthologues are present in all corynebacterineae. Whereas the Rv3807 gene apparently is not essential, contrary to all the other genes related to decaprenyl-phospho-arabinose synthesis annotated as essential genes [14]. In this study, we expressed and identified the Rv3807c gene in Escherichia coli strain and predicted the structure and characteristics of Rv3807c protein by bioinformatics analysis.
Materials and Methods
Amplification of Rv3807c Gene by PCR
The nucleotide sequence of M. tuberculosis Rv3807c was obtained from Tuber Gene Data and was as a template for designing primers using Primer premier 5.0 software. The forward primer 5′-ATTCATATGGTGGCCGTGCAGTCGGC-3′ (NdeI) and the backward primer 5′-ATTGGATCCTCATCTCTTCCGGGCCCTTTGC-3′ (BamHI). NdeI and BamHI endonuclease restriction sites were included at the 5′-end of the primers respectively. PCR was performed in a final volume of 50 μl. The PCR conditions consisted of pre-denaturation (94 °C, 5 min) and 30 cycles of amplification including denaturation (94 °C, 30 s), annealing (62 °C, 30 s) and primer extension (72 °C, 1 min). After the last amplification cycle there was a final extension at 72 °C for 10 min. All of the PCR product was separated by 1 % agarose gel and purified with TIANgel midipurification kit (TIANGEN, China).
Cloning and Expression of Rv3807c Gene
The purified PCR product of Rv3807c was ligated to pMD18-T vector (TAKARA, China) by using the T4 DNA ligase overnight at 16 °C. 10 μl ligation mixtures were transformed into the Novablue-competent cells (Novagen, America). The positive recombinant plasmid pMD18-Rv3807c was confirmed by restriction endonuclease digestion and DNA sequencing. The pMD18-Rv3807c was digested with NdeI and BamHI and the Rv3807c was subcloned into the NdeI and BamHI sites of pColdII (Takara, Japan) to yield an expression plasmid pColdII-Rv3807c. The pColdII-Rv3807c was transformed into the competent E. coli BL21 (DE3) (Novegen, America). The E. coli BL21(DE3) cells harboring the pColdII-Rv3807c plasmid were cultured in 50 ml LB medium supplemented with ampicillin (100 μg/ml) at 37 °C until A600 reaching 0.5–0.6. The cell cultures were induced with isopropyl-d-thiogalactopyranoside (IPTG) at a final concentration of 1 mM at 16 °C for 4 h. The bacterial cells were harvested, resuspended in the cell lysis buffer solution (50 mM Tris–HCl, 1 mM EDTA, 100 mM NaCl, 25 mM MgCl2 and 5 % glycerol), and disrupted by sonication on ice. The lysate mixture was centrifuged at 20,000×g for 30 min to separate the supernatant and pellet. To obtain the membrane protein, the supernatant was ultracentrifuged at 200,000×g for 1 h, and the membrane-enriched pellet was harvested and homogenized in 1 ml cell-lysis buffer.
Analysis of Rv3807c Protein by Western Blotting
SDS-PAGE was run using 15 % gel. Samples from the acrylamide-gel were transferred onto a nitrocellulose membrane under semidry transfer condition for Western blotting. Primary and secondary antibodies of monoclonal anti-polyhistidine (Sigma, America) and Horse-anti-mouse-IgG conjugated with alkaline phosphatase (ZSGB-Bio, China) were respectively applied according to standard Western blotting protocol. The membrane was stained in the solution mixture of BCIP/NBT (ZSGB-Bio, China) and visualized in the dark condition for 5 min.
Purification of Rv3807c Protein
The recombinant pColdII-Rv3807c protein was purified using Ni-NTA affinity chromatography columns (Qiagen, Germany). The purification was conducted according to manufacture’s protocol. The purified protein was subsequently evaluated on SDS-PAGE and identified via Western-blotting.
Bioinformatics Analysis of Rv3807c Protein
The amino acid sequences of Rv3807c were retrieved from the tuberculist in Genolist Genome Browser (http://www.ncbi.nlm.nih.gov/). The programs of PredictProtein (http://www.predictprotein.org/) and ProtParam (http://web.expasy.org/protparam/) were used to analyze the basic physical and chemical parameters. For the subcellular location, cleavage site and signal peptides prediction, TargetP (http://www.cbs.dtu.dk/services/TargetP/), PSORT (http://www.psort.org/), TMHMM (http://www.cbs.dtu.dk/services/TMHMM-2.0/) and SignalP (http://www.cbs.dtu.dk/services/SignalP/)programs were employed. The information on homology sequences and Prosite were obtained via InterProScan (http://www.ebi.ac.uk/Tools/pfa/iprscan/), BLASTP (http://npsa-pbil.ibcp.fr/) and PredictProtein. Secondary structure and comparative protein structure model were computed using the SOSUI (http://bp.nuap.nagoya-u.ac.jp/sosui/), PredictProtein, COILS (http://embnet.vital-it.ch/software/COILS_form.html), SOPMA (http://npsa-pbil.ibcp.fr/) and ModBase software (http://modbase.compbio.ucsf.edu/).
Results
Amplification of Rv3807c Gene by PCR
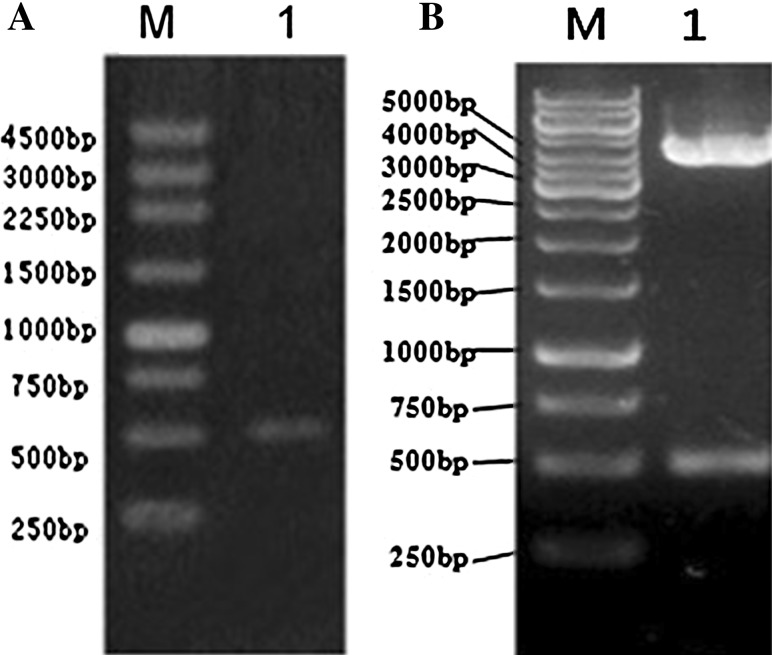
Amplification product of Rv3807c gene by PCR using the template of M. tuberculosis H37Rv genomic DNA and high fidelity LA Taq-DNApolymerase generated an expected 507 bp DNA fragment band(containing restriction site linker) as shown on the 1 % agarose gel under UV (Figure 1a).
Fig. 1.
PCR amplification of Rv3807c gene and the identification of recombinant plasmid pColdII-Rv3807c by endonuclease restriction analysis a Rv3807c gene amplified from Mycobacterium tuberculosis was shown on agarose gel. Lane M 250 bp DNA ladder, Lane 1 PCR product of Rv3807c gene; b recombinant plasmid pColdII-Rv3807c was identified via restriction analysis. Lane M 1 Kb DNA ladder; Lane 1 pColdII-Rv3807c gene digested by Nde I0/BamH I
Cloning and Expression of Rv3807c Gene
The Rv3807c gene was cloned into the multiple cloning site region of pColdII expression vector to form the recombinant plasmid separately. The identified plasmid was transformed into an expression strain. In early experiments, the Rv3807c gene was ligated to pET16b vector, and the plasmid of pET16b-Rv3807c was then transformed into the E. coli Er2566 well expressing membrane proteins. However, less supernatant target protein was produced than inclusion bodies formation in the pellet. The pColdII vector was chosen for expression finally.
After the recombinant plasmids, pColdII-Rv3807c, transformed into the E. coli BL21 (DE3) for expressing, correct orientation was verified with the aid of the restriction NdeI/BamHI enzymes. Figure 1b shows the expected size of DNA fragments of 4,369 bp/500 bp on agarose gel, which are consistent with the length deposited in Clone manager (Fig. 1b).
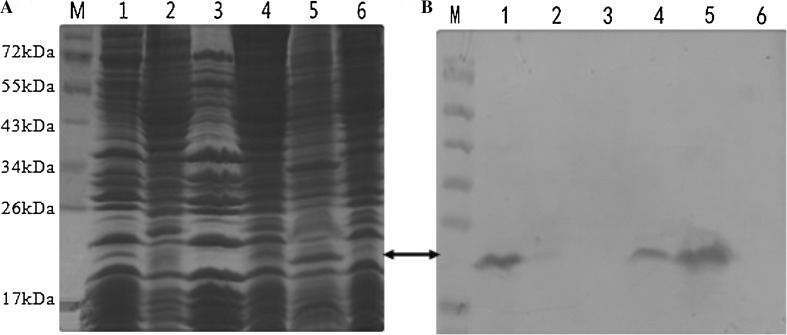
The 6× His-Rv3807c protein also showed an expected strong 20 kDa protein band on SDS-PAGE. Western blot analysis of total proteins demonstrated the observed protein band reacting with the specific antibodies and the protein mainly in the insoluble fraction yet (Figure 2a, b). The purity of the recombinant protein pColdII-Rv3807c was revealed no expected band near 20 kDa on the nitrocellulose membrane by western blotting.
Fig. 2.
SDS-PAGE analysis and Western blotting identification of pColdII-Rv3807c total proteins expressed in E. coli BL21 (DE3) a SDS-PAGE and b Western-blot analysis of pColdII-Rv3807c total proteins expressed. Lane M protein molecular mass standards; Lane 1 pColdII-Rv3807c membrane protein, the pellet after ultracentrifuged; Lane 2 the supernatant after ultracentrifuged; Lane 3 negative control, pColdII membrane protein; Lanes 4, 5 pColdII-Rv3807c supernatant and pellet proteins induced by 1 mM IPTG for 4 h; Lane 6 negative control, pColdII supernatant protein induced by 1 mM IPTG for 4 h
Bioinformatics Analysis of Characteristics and Structures of Rv3807c Protein
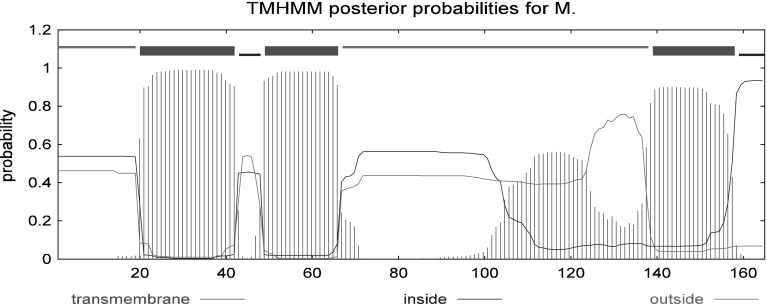
The Rv3807c protein containing 165 amino acids with an estimated molecular mass of 17.2 kDa and an isoelectric point 12.29 had a signal peptide near the N-terminal and three transmembrane regions (Fig. 3). It was predicted as a cytoplasmic membrane protein by PSORT program. The amino acid sequence of Rv3807c from M. tuberculosis H37Rv showed a similar core structure with phosphatidic acid phosphatase type 2/haloperoxidase, with examples as family PAP2 (PF01569), acid phosphatase homologues (SM00014) and acid phophatase/vanadium-dependent haloperoxidase (SSF48317), as shown in Fig. 4. Secondary structure analysis revealed that Rv3807c protein contained 49.7 % α-helix, 3.03 % β-turn, 13.33 % extended strand and 33.94 % random coil, containing four transmembrane helices (Fig. 5).
Fig. 3.
Prediction of subcellular location and transmembrane regions of Rv3807c protein
Fig. 4.

The acid phosphatase homology sequences (51-153) predicted according to the interpromember databases used to construct an entry
Fig. 5.

Secondary structure of Rv3807c protein, h alpha helix, t beta turn, e extended strand, c random coil
Discussion
Decaprenyl-phospho-d-arabinose (DPA) is the only known donor of polymerized Araf residues for AG biosynthesis in mycobacteria and is essential for the biosynthesis of the cell wall in M. tuberculosis [5]. The initial formation of DPA is followed three reactions: transfer of a 5′-phosphopentose to DP, removal of the 5′-phosphate, and epimerization [12]. The enzymes of Rv3806c, Rv3790/Rv3791 have been reported to be involved in catalyzing the first and final steps, which are essential for the cell wall in Mycobacterium. However, the phospholipid phosphatase involved has not been identified yet. The presence of an unknown PAP2-family phospholipid phosphatase (Rv3807c) located next to the phosphoribosyltransferase (Rv3806c) was predicted to catalyze the reaction of dephosphorylation [11]. In this study, we constructed the recombinant plasmid pColdII-Rv3807c to clone and express the Rv3807c gene and identified the localization, functional and structural characteristics of the Rv3807c protein to enable further confirmatory study on the relationship between Rv3807c and DPA biosynthesis.
The Rv3807c protein was successfully expressed in E. coli BL21 (DE3) after PCR amplification. The expressed protein was detected in the membrane proteins fraction upon centrifugation and Western blotting analysis. The purification of Rv3807c protein from the supernatant protein was unsuccessful. The hydrophobic interaction and the folding characteristics of the Rv3807c N-terminal protein may be the possible reasons to have interfered with the successful purification. In our experiment, Rv3807c–f, the DNA fragment of Rv3807c gene, was estimated to be the soluble part of the Rv3807c N-terminal protein, and was also cloned and expressed using the same methods as Rv3807c gene. The purified product of Rv3807c–f protein with Ni-NTA column revealed the expected protein band near 17 kDa on the nitrocellulose membrane ultimately (data not shown). The functional characterization of the purified Rv3807c–f protein is ongoing in our laboratory, and this can provide new evidence for the functional studies of Rv3807c protein.
To begin with, genomic information, in combination with bioinformatics provides sufficient detail into functional characterization of processes encoded in the M. tuberculosis genome, and the next step is experimental verification of gene function, which is particularly important for further characterization of metabolic pathways [15]. The Rv3807c protein was found in the membrane fraction and is consistent with the prediction of three transmembrane domains by bioinformatics analysis, in which the N-terminal peptide was found located outside of the cytoplasmic membrane. Additionally, functional and structural characteristics were predicted to be similar with phosphatidic acid phosphatase type2/haloperoxidase, which catalyze the dephosphorylation of phosphatidate [16]. Therefore the Rv3807c is considered to be the potential and suitable candidate for the phospholipid phosphatase in DPA biosynthesis and with further investigations could be a new potent target for antituberculosis drugs.
In summary, the proposed pathway of DPA is unique in M. tuberculosis and represents a possible target for new drug discovery along with the emergence of multi-resistant strains recently. Ethambutol, a first-line drug for treatment of TB with pleiotropic effects, however, does not block the synthesis of DPA [11], which directly influence AG biosynthesis, even the formation of the cell wall in mycobacterium [10, 17]. It remains to be determined whether the speculated enzyme Rv3807c catalyses the conversion of DPPR to DPR and its precise role in the biosynthesis of DPA.
Acknowledgments
This study is supported by the grants from National Natural Science Foundation of China (30970647).
Conflict of interest
The authors declare that they have no conflicts of interest.
Abbreviation
- BCIP
5-Bromo-4-chloro-3-indolylphosphate
- DP
Decaprenyl-phosphate
- DPA
Decaprenyl-phospho-d-arabinose
- DPPR
Decaprenylphosphoryl-d-5-phosphoribose
- DPR
Decaprenylphosphoryl-d-ribose
- NBT
Nitroblue tetrazolium
- Ni-NTA
Ni-nitrilotriacetic acid
- EDTA
Ethylenediaminetetraacetic acid
- PAGE
Polyacrylamide gel electrophoresis
- PRPP
5-Phosphoribose-1-diphosphate
- SDS
Sodium dodecyl sulfate
References
- 1.Brennan PJ. Structure, function, and biogenesis of the cell wall of Mycobacterium tuberculosis. Tuberculosis (Edinb) 2003;83(1–3):91–97. doi: 10.1016/S1472-9792(02)00089-6. [DOI] [PubMed] [Google Scholar]
- 2.Crick DC, Schulbach MC, Zink EE, Macchia M, Barontini S, Besra GS, et al. Polyprenyl phosphate biosynthesis in Mycobacterium tuberculosis and Mycobacterium smegmatis. J Bacteriol. 2000;182(20):5771–5778. doi: 10.1128/JB.182.20.5771-5778.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crick DC, Mahapatra S, Brennan PJ. Biosynthesis of the arabinogalactan–peptidoglycan complex of Mycobacterium tuberculosis. Glycobiology. 2001;11(9):107R–118R. doi: 10.1093/glycob/11.9.107R. [DOI] [PubMed] [Google Scholar]
- 4.Scherman M, Weston A, Duncan K, Whittington A, Upton R, Deng L, et al. Biosynthetic origin of mycobacterial cell wall arabinosyl residues. J Bacteriol. 1995;177(24):7125–7130. doi: 10.1128/jb.177.24.7125-7130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wolucka BA, McNeil MR, de Hoffmann E, Chojnacki T, Brennan PJ. Recognition of the lipid intermediate for arabinogalactan/arabinomannan biosynthesis and its relation to the mode of action of ethambutol on mycobacteria. J Biol Chem. 1994;269(37):23328–23335. [PubMed] [Google Scholar]
- 6.Crellin PK, Brammananth R, Coppel RL. Decaprenylphosphoryl-beta-d-ribose 2′-epimerase, the target of benzothiazinones and dinitrobenzamides, is an essential enzyme in Mycobacterium smegmatis. PLoS One. 2011;6(2):e16869. doi: 10.1371/journal.pone.0016869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horita Y, Takii T, Kuroishi R, Chiba T, Ogawa K, Kremer L, et al. Synthesis and evaluation of anti-tubercular activity of new dithiocarbamate sugar derivatives. Bioorg Med Chem Lett. 2011;21(3):899–903. doi: 10.1016/j.bmcl.2010.12.084. [DOI] [PubMed] [Google Scholar]
- 8.Buroni S, Pasca MR, de Jesus Lopes Ribeiro AL, Degiacomi G, Molteni E, Riccardi G. Antituberculars which target decaprenylphosphoryl-β-d-ribofuranose 2′-oxidase DprE1: state of art. Appl Microbiol Biotechnol. 2012;94(4):907–916. doi: 10.1007/s00253-012-4013-4. [DOI] [PubMed] [Google Scholar]
- 9.Huang H, Scherman MS, D’Haeze W, Vereecke D, Holsters M, Crick DC, et al. Identification and active expression of the Mycobacterium tuberculosis gene encoding 5-phospho-α-d-ribose-1-diphosphate: decaprenyl-phosphate 5-phosphoribosyltransferase, the first enzyme committed to decaprenylphosphoryl-d-arabinose synthesis. J Biol Chem. 2005;280(26):24539–24543. doi: 10.1074/jbc.M504068200. [DOI] [PubMed] [Google Scholar]
- 10.Jiang T, He L, Zhan Y, Zang S, Ma Y, Zhao X, et al. The effect of MSMEG_6402 gene disruption on the cell wall structure of Mycobacterium smegmatis. Microb Pathog. 2011;51(3):156–160. doi: 10.1016/j.micpath.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 11.Wolucka BA. Biosynthesis of d-arabinose in mycobacteria—a novel bacterial pathway with implications for antimycobacterial therapy. FEBS J. 2008;275(11):2691–2711. doi: 10.1111/j.1742-4658.2008.06395.x. [DOI] [PubMed] [Google Scholar]
- 12.Mikusova K, Huang H, Yagi T, Holsters M, Vereecke D, D’Haeze W, et al. Decaprenylphosphoryl arabinofuranose, the donor of the d-arabinofuranosyl residues of mycobacterial arabinan, is formed via a two-step epimerization of decaprenylphosphoryl ribose. J Bacteriol. 2005;187(23):8020–8025. doi: 10.1128/JB.187.23.8020-8025.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eoh H, Brown AC, Buetow L, Hunter WN, Parish T, Kaur D, et al. Characterization of the Mycobacterium tuberculosis 4-diphosphocytidyl-2-C-methyl-d-erythritol synthase: potential for drug development. J Bacteriol. 2007;189(24):8922–8927. doi: 10.1128/JB.00925-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sassetti CM, Boyd DH, Rubin EJ. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol. 2003;48(1):77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 15.Slayden RA, Jackson M, Zucker J, Ramirez MV, Dawson CC, Crew R, et al. Updating and curating metabolic pathways of TB. Tuberculosis (Edinb) 2013;93(1):47–59. doi: 10.1016/j.tube.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carman GM, Han GS. Roles of phosphatidate phosphatase enzymes in lipid metabolism. Trends Biochem Sci. 2006;31(12):694–699. doi: 10.1016/j.tibs.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dong X, Bhamidi S, Scherman M, Xin Y, McNeil MR. Development of a quantitative assay for mycobacterial endogenous arabinase and ensuing studies of arabinase levels and arabinan metabolism in Mycobacterium smegmatis. Appl Environ Microbiol. 2006;72(4):2601–2605. doi: 10.1128/AEM.72.4.2601-2605.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]