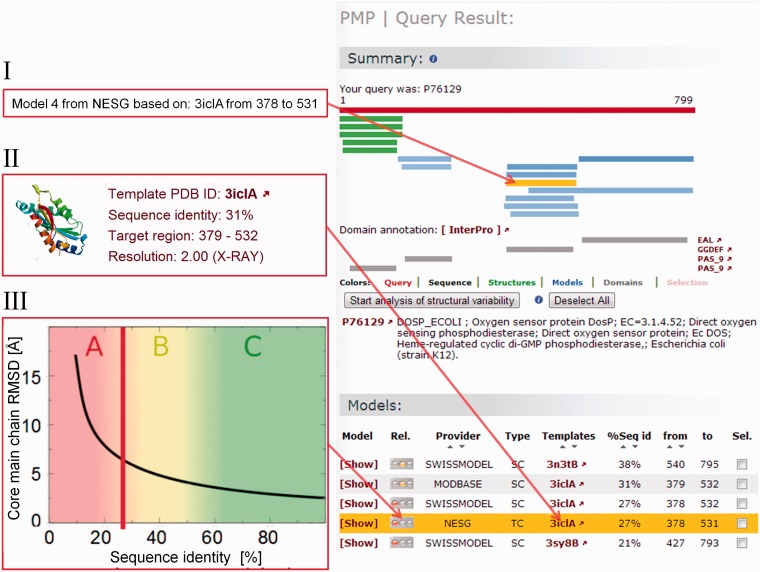
Figure 2.
PMP results summary page for the oxygen sensor protein (UniProt Accession Code P76129). Experimentally determined structures for the protein (i.e. PDB entries with sequence identity >90%) are shown in green, models in shades of blue, the darker the higher the sequence identity of the template to the target protein. Pfam domain annotations are depicted in gray, to the right of each domain is a direct link to InterPro providing more information about the different domains. Apart from the graphical representation of structural coverage, detailed lists of experimental structures (not shown) and models are presented. As example, a model from NESG, which was built on the template 3ICL chain A, is highlighted in orange (mouseover text of corresponding model bar is shown in panel I). More details of the template used is given in panel II, where also the template deposition date, a preview image of the structure and the experimental method are given. Expected quality of the model based on the sequence identity between aligned target and template sequences is illustrated in panel III, where a vertical red bar marks the sequence identity of 27% to the template and the expected model accuracy based on the work of Chothia and Lesk (37).