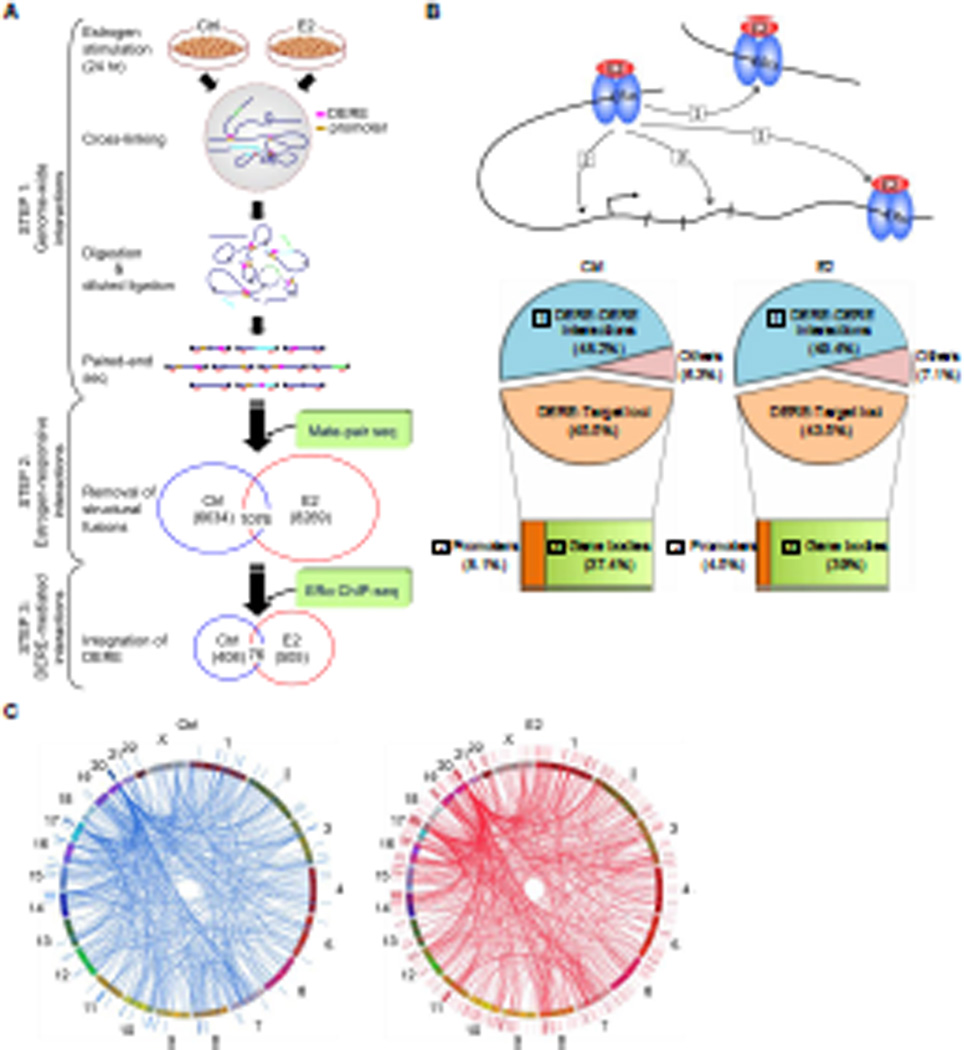
Figure 1. Integrative Mapping of ERα-mediated Chromatin Interactions Based on Next-generation Sequencing Approaches.
(A) Integrative scheme of identifying ERα/DERE-mediated chromatin interaction sites in E2-stimulated MCF-7 human breast cancer cells. Chromosome conformation capture (3C) assay coupled with paired-end sequencing was performed on both untreated (Ctrl) and estrogen-treated (E2, 70 nM) MCF-7 cells to survey chromatin interaction events in a genome-wide manner (STEP 1) (see also Figure S1A–B and Tables S1–2). To identify genuine interaction sites, genomic fusions and self-ligated fragments mapped by mate-pair sequencing were filtered-out from the 3C-seq dataset (STEP 2) (see also Tables S3–6). The filtered data were then integrated with ERα ChIP-seq datasets (0 and 24 hr, respectively) and distant estrogen response elements (DEREs) were mapped to define DERE-associated chromatin interaction events (STEP 3) (see also Tables S7).
(B) Genomic distribution of ERα-mediated chromatin interaction sites. ERα-mediated interaction sites mapped within 10-kb regions of DEREs, which have no known target genes, were defined as DERE-DERE interactions. In the target loci category, the regions within 10-kb upstream and 1-kb downstream of the transcription start site (TSS) of a gene were defined as promoters. “Others” were defined as ERα-mediated interaction sites mapped in gene-desert regions.
(C) Circular visualization of ERα-mediated interactions upon E2 stimulation. Circular plots depict interactive loci of different chromatin loops using the Circos software (http://mkweb,bcgsc.ca/circos/). Chromosomes are individually colored. The locations of DEREs are represented as lines outside the chromosomes “circle”. Four clustered DEREs were identified in 1p13, 3p14, 17q23, and 20q13 regions (see also Figure S1C).