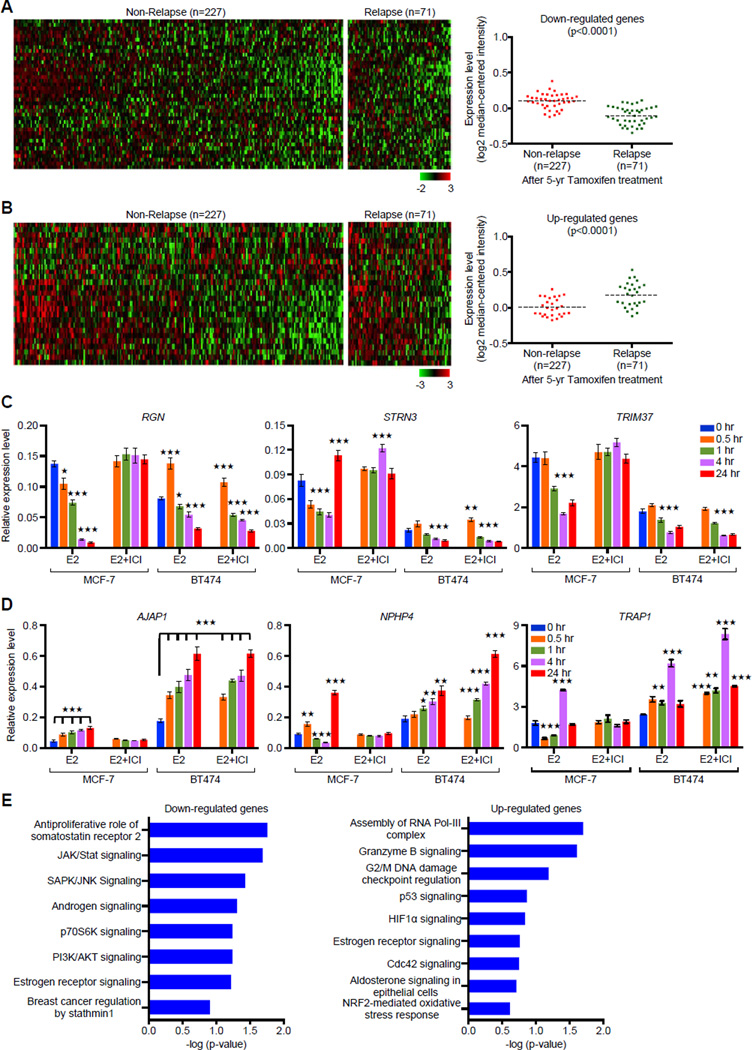
Figure 8. Expression Signature of DERE-interacting Genes Correlates with Relapse after Tamoxifen Therapy.
(A–B) In silico analysis of DERE-regulated genes in an ERα-positive breast tumor cohort is associated with relapse. A breast cancer cohort including a total of 298 patients with ERα-positive breast tumors and 5-year tamoxifen treatment was used to study the clinical significance of DERE-regulated genes (Symmans 2010). Forty down-regulated (A) and twenty-seven up-regulated (B) genes were significantly associated with relapse after tamoxifen treatment (p<0.0001) (see also Figure S7A–B for validation analysis using another independent cohort within endocrine therapy history).
(C–D) Expression analysis of 26 DERE-interacting genes Involved in tamoxifen resistance. Quantitative RT-PCR analysis was performed on MCF-7 and BT474 cells treated with E2 (70 nM) alone/and ERα antagonist- ICI 182,780 (ICI, 1 µM) in five time periods (0, 0.5, 1, 4, and 24 hr). Expression data were shown in 26 bar charts with individual genes (see also Figure S7C–D). Mean ± SD (n=6 replicates in two biological batches). (C) Down-regulated genes; (D) Up-regulated genes.
(E) Signaling pathways associated with DERE-regulated genes in tamoxifen resistance. Ingenuity Pathway Analysis was used to determine signaling pathways associated with down- and up-regulated genes (identified from A and B) in tamoxifen resistance. Mean ± SD (n=6 replicates in two batches of treatment). ***, p<0.001; **, p<0.01; *, p<0.05 (Student’s t test), comparing to control cells (time point “0 hr”).