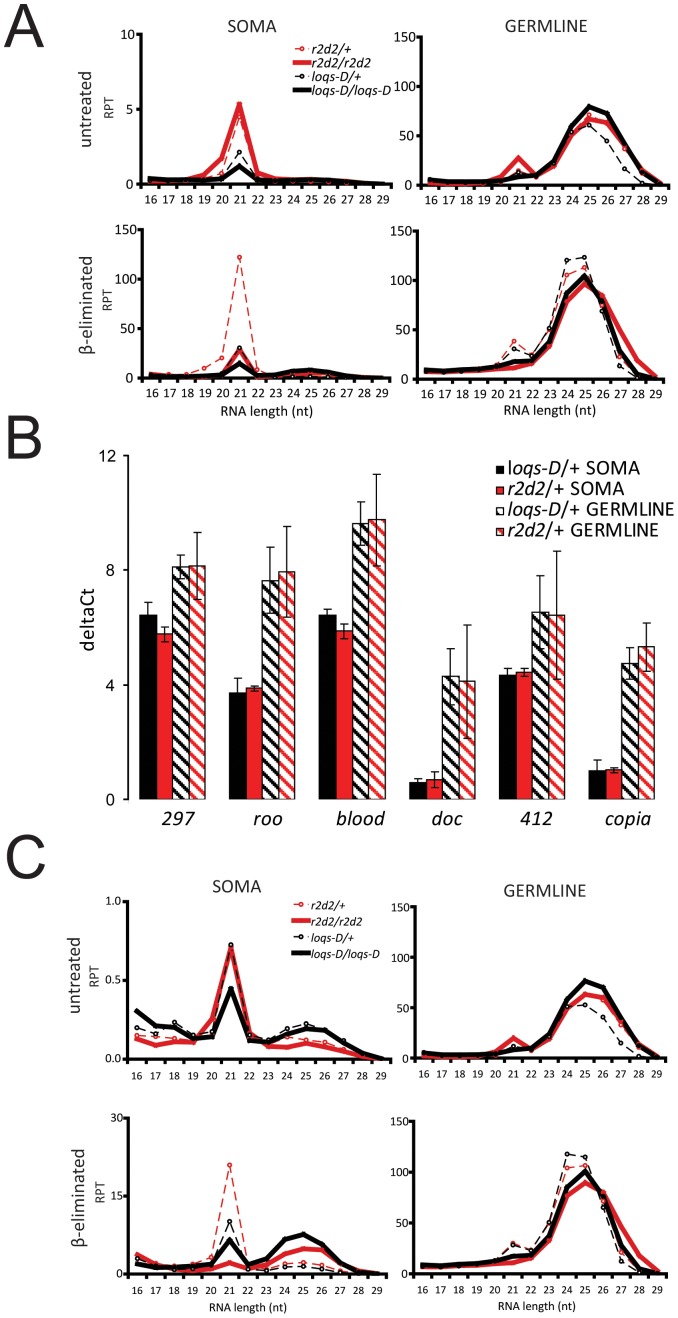
Figure 1. Length distribution of transposon-matching small RNAs identified in this study.
A) Reads of each library originating from soma and germline were mapped to the reference containing a transposon sequence collection. Transposon matching small RNAs were analyzed for their size distribution and normalized to total genome matching reads. The normalized counts were expressed as reads per thousand (RPT). B) The steady state transcript levels of 297, TNFB, roo and blood transposable elements were examined by qRT-PCR. RNA was isolated from three biological replicates of heterozygous loqs-D and r2d2 mutants separated in somatic and germline tissue, respectively. The doc, 412 and copia transposons were included for comparison. Ct-values for each transposon were normalized to the rp49 control (delta Ct). Values are mean ± SD (n = 3). C) The length distribution of transposon matching small RNAs in r2d2 and loqs-D mutants after exclusion of roo, 297, TNFB and blood transposons.