Figure 7.
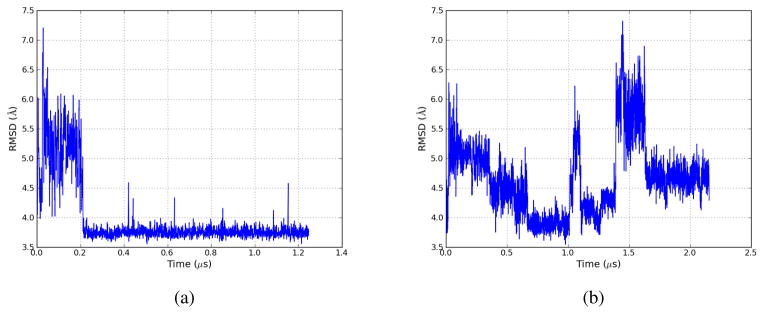
RMSD plots for two Villin NLE Amber99-SB, GB-OBC implicit-solvent simulations run with LTMD. RMSD was calculated using Cα atoms, excluding the first two and last two residues, against the folded structure. The minimum RMSDs of each simulation are 3.6 Å and 3.5 Å, respectively. Figure 7b shows multiple folding and unfolding events, occurring approximately every 0.5 μs.
