Figure 1.
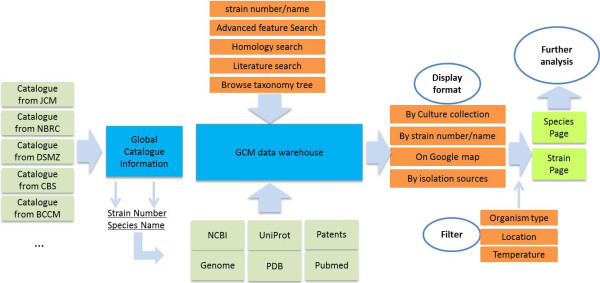
Scheme of the workflow of GCM. Catalogue information from each of collection, shown on the left, is used to construct the framework of the global catalogue database. Species name and strain numbers are collected from the catalogue information, and are further used to identify and extract information from public database such as Genbank, NCBI Genome, SwissProt, PDB, Pubmed and the patent database. The data warehouse is built in a SQL database, and can be accessed via a web interface through different search options. Search results may be displayed in different formats allowing users to refine the results by using filters. The final results are displayed either as a strain page or they can be gathered into a species page depending on the query. BLAST and ClustalW are provided for further analysis of the results.
