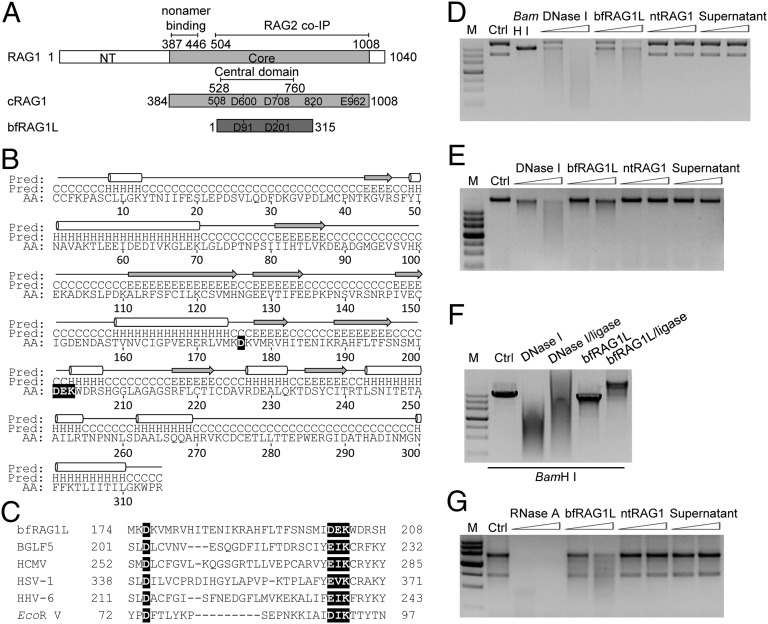
Fig. 1.
The structural and functional analyses of bfRAG1L. (A) Alignment between RAG1 and bfRAG1L. The conserved DDE motif residues are indicated in the box. Numbers represent positions of amino acids. The nonamer-binding domain, RAG2 binding domain, and the central domain are marked with straight lines. NT, N-terminal. Core, the core region of mouse RAG1. (B) The predicted secondary structure is labeled with cylinders (α-helices) and arrows (β-strands). (C) Sequences alignment of the conserved nuclease active motif (highlighted in black) in bfRAG1L, several herpes virus nucleases, and EcoR V. Nuclease activity assays were performed (SI Appendix). The nuclease activities of bfRAG1L (0.1 μg or 0.2 μg) on the substrates plasmid pBR322 (D), genomic DNA (E), or RNA from HEK-293T cells (G) are shown. The incubations with substrates DNA are executed for 8 h, or with substrate RNA for 3 h. Ctrl, represents respective untreated substrate; M, DNA marker. (F) The religation products of BamH I-digested linear plasmid pBR322 degraded by 0.1 μg of bfRAG1L or DNase I. Ctrl, untreated BamH I-digested linear plasmid pBR322. Supernatant, purified bfRAG1L protein immunodepleted with anti-bfRAG1L serum. The results are representative of three independent experiments.