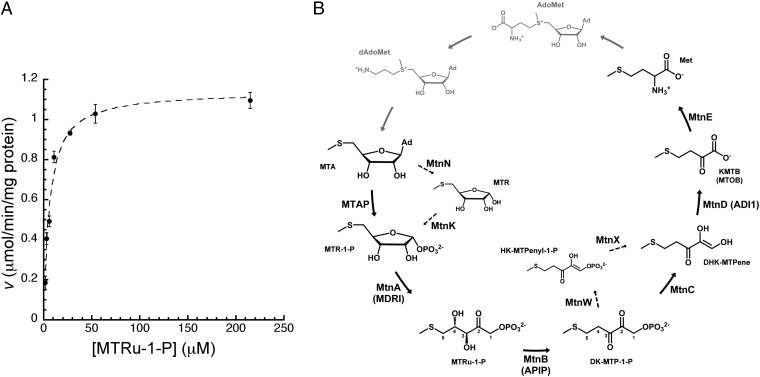
Fig. 1.
APIP as an MtnB enzyme in the methionine salvage pathway. (A) Initial reaction rate was plotted at seven different concentrations of the substrate MTRu-1-P for Michaelis-Menten kinetic analysis. Data represent mean values with SE from three independent measurements. (B) Methionine salvage pathway characterized in Homo sapiens and Saccharomyces cerevisiae converts MTA to methionine (Met) through the common six enzymatic reactions. Dashed line represents B. subtilis methionine salvage reaction steps distinct from H. sapiens and S. cerevisiae. Gray colored enzymatic steps and metabolites represent biochemical links that are not conceptually part of the methionine salvage pathway. AdoMet, S-adenosyl-l-methionine; dAdoMet, decarboxylated AdoMet; DHK-MTPene, 1,2-dihydroxy-3-keto-5-methylthiopentene; HK-MTPenyl-1-P, 2-hydroxy-3-keto-5-methylthiopentenyl-1-phosphate; Met, l-methionine; MTOB, 4-methylthio-2-oxobutyrate; MTR, 5-methylthioribose.