Fig. 1.
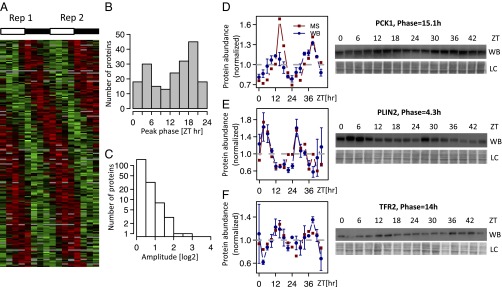
SILAC-based MS identifies the diurnal proteome in mouse liver. (A) Heatmap showing the rhythmic proteome (n = 195, FDR < 0.25); 2 independent d (each comprising eight time points) are juxtaposed; data are standardized by rows, and gray blocks indicate missing data. (B and C) Phase and peak-to-trough amplitude (log2 units) distribution for the rhythmic proteins (FDR < 0.25). (D–F) Examples of rhythmic proteins encoded by the individual genes (D) Phosphoenolpyruvate CarboxyKinase 1 (PCK1; P = 1.3e-05, q = 0.0060), (E) PeriLipIN2 (PLIN2; P = 0.00083, q = 0.068), and (F) TransFerrin Receptor protein 2 (TFR2; P = 6.39e-06, q = 0.0036) confirmed by Western blot (WB) analysis (data are normalized to the temporal mean). The two biological replicates are shown as ZT0–ZT21 (replica 1) and ZT24–ZT45 (replica 2). For the WBs, 16 time points show the mean and SE from two independent biological samples. Naphtol blue-black staining of the membranes was used as a loading control and serves as a reference for normalization of the quantified values. The ZTs at which the animals were killed are indicated. LC, loading control.
