Abstract
The presence and conservation of GnRH2 across vertebrate species suggest important biological roles. However, the function of GnRH2 remains unclear. A good research model for GnRH2 functional studies is still lacking largely due to the absence of GnRH2 in the widely used mouse model. Hence, we used the zebrafish, for which powerful genetic tools are available, and developed a transgenic (Tg) line expressing enhanced green fluorescence protein (eGFP). The high sensitivity of eGFP, which can diffuse throughout the neuron, enables us to document the complete projectome of GnRH2 neurons at different developmental stages. Fine projection structures were observed without sacrificing the fish. Crossed with the GnRH3:tdTomato Tg line, the GnRH2:eGFP Tg line provides us with an opportunity to visualize the entire GnRH system simultaneously in one organism. This work will provide a framework to understand the function of the highly-conserved GnRH2 system.
Since the discovery of the first gonadotropin-releasing hormone (GnRH) decapeptide (referred to as GnRH1) in the mammalian hypothalamus1, diverse forms have been found in vertebrates2 and even in invertebrates3,4,5, indicating an ancient evolutionary origin. Three known distinct neuronal populations of GnRH exist in the vertebrate brain: the forebrain population (GnRH1), the midbrain tegmentum population (GnRH2), and the hypothalamic-preoptic area region subset that expresses teleost fish-specific GnRH36,7. Some teleost fish have lost either the GnRH1 or GnRH3 gene, and the remaining form will take over the non-redundant functions of the lost gene7,8. GnRH1's functions are relatively well known as the central initiator of the reproductive endocrine axis through stimulating the biosynthesis and release of the pituitary gonadotropins luteinizing hormone (LH) and follicle-stimulating hormone (FSH). GnRH2 is the only GnRH isoform that is completely conserved in almost every extant vertebrate species9 and is exclusively expressed in the neurons of the dorsal synencephalon/rostral tegmentum area in most vertebrate species10. In contrast to the vast knowledge regarding the hypothalamic GnRH neuronal system (either GnRH1 or GnRH3 depending on the species), very little is known about the biology and functions of GnRH2. This gap in understanding probably reflects the fact that a GnRH2 system is lacking in many widely-used mammalian research models, including mouse11.
Determining the anatomical, biochemical and functional organization of the nervous system is an important task of neurobiology. A good visualization of GnRH2 cell bodies and their projections will contribute to our understanding of the GnRH2 system. Even though GnRH2 expression has been detected in multiple normal12,13,14,15,16 and malignant tissues/sites17,18,19,20,21,22 and has been characterized during different biological processes23,24,25, a clear description of GnRH2 neuronal groups and their local and far-ranging projections is still lacking. To accomplish this objective, we chose zebrafish as our research model. Besides the similar 2-GnRH system that zebrafish shares with primates and humans, it also has various research advantages, such as translucent embryos, a relatively well established functional nervous system at about 4–5 days post-fertilization (dpf) and clear visibility at a cellular resolution level26 due to its small size. We tagged zebrafish GnRH2 neurons with enhanced green fluorescence protein (eGFP) under the control of the cis-regulatory region from the zebrafish GnRH2 gene-containing BAC clone (DKEYP-87D8) and have been able to generate a large quantity of high resolution in vivo data describing the soma and projection behavior and distribution of GnRH2 neurons. Furthermore, the crossing of GnRH2:eGFP Tg fish with the GnRH3:tdTomato Tg line provides us with a detailed delineation of the projections of the entire zebrafish GnRH system.
Results
Genetic labeling of GnRH2 neurons using modified BAC DNA
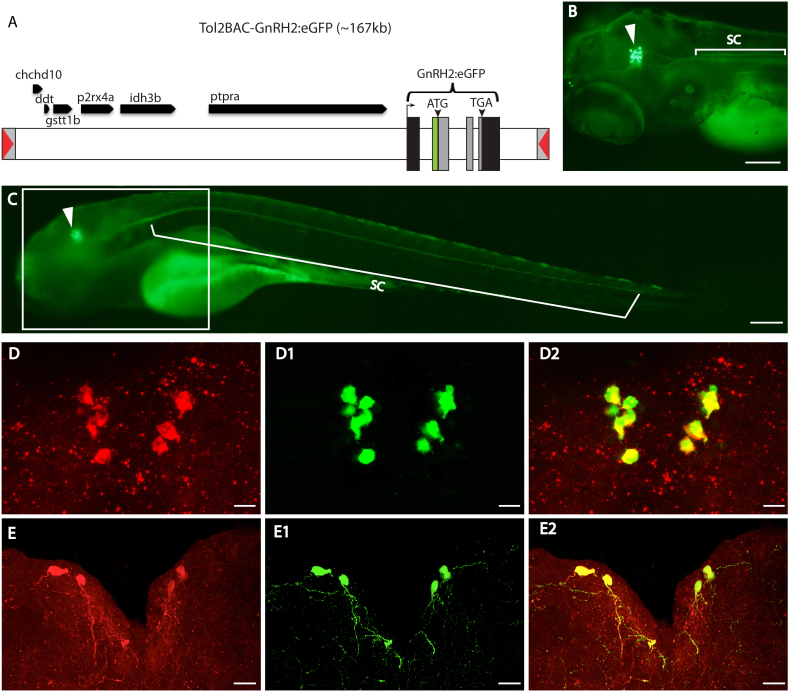
To our knowledge, no fluorescence-labeled GnRH2 transgenic zebrafish has been generated thus far. After failing to faithfully mimic endogenous GnRH2 expression using various lengths of flanking sequences upstream of GnRH2 (ranging from 2 kb to 12 kb), we used BAC DNA containing a 130 kb upstream region with the GnRH2 coding sequence replaced with eGFP (Figure 1A). Stable ZF lines exhibiting strong and specific eGFP expression patterns (Figures 1B, C) were generated, and the midbrain tegmentum expression of GnRH2:eGFP BAC was shown to be specific for GnRH2 as corroborated by the perfect overlapping between the eGFP positive cells and the GnRH2 mRNA (in situ hybridization) and protein (immunocytochemistry) expressing cells in the tegmentum at different developmental stages (Figures 1D–E2 and S1A–B2). The specificity of the antibody used for immunocytochemistry (ICC) was confirmed through the abolishment of the ICC signal following pre-absorption of the GnRH2 antibody with excess recombinant GnRH2-GAP protein, against which the GnRH2 antibody was generated (Figures S1B–B2). Although the GnRH2 antibody can recognize the GnRH2 cells (Figure 1D) and fibers (Figure 1E–E2), the immunostaining procedure cannot efficiently label the fibers (Figures S1A–A2) due to its poor signal-to-noise ratio, compared with endogenous eGFP signals. This technical shortfall leads to some non-specific background in the ICC27, (Figure 1D), which makes identifying GnRH2 projections through ICC a challenge, and thereby favors the use of the eGFP approach for monitoring the GnRH2 system with a much higher resolution.
Figure 1. Generation of GnRH2:eGFP Tg zebrafish.
(A) Schematic diagram of modified BAC (DKEYP-87D8) DNA used to generate GnRH2:eGFP Tg zebrafish. GnRH2 exons are labeled as black and grey vertical bars. The approximate sizes of the exons/introns and other genes are represented by the bar lengths. The grey boxes represent the genomic DNA encoding the entire GnRH2 open reading frame (ORF) that was replaced by eGFP. The inverted red triangles represent iTol2 elements. Dorsal left view (B) and lateral view (C) of a 4 dpf GnRH2:eGFP Tg larva (anterior to the left). Strong expression, indicated by green fluorescence, is seen in neuronal cell bodies in the midbrain tegmentum area (arrowhead) and extending continuously into the spinal cord. ICC staining (D & E) using GnRH2 antibody (red) with green fluorescent signals from endogenously expressed eGFP proteins (D1 & E1). Merged views (D2 & E2) of 11 dpf and adult GnRH2:eGFP Tg brain sections (z-stack from 100 μm cross-sections, with dorsal at the top), showing co-localization of eGFP signal in GnRH2 neurons. Scale bars: 100 μm (B & C) and 20 μm (D–E2).
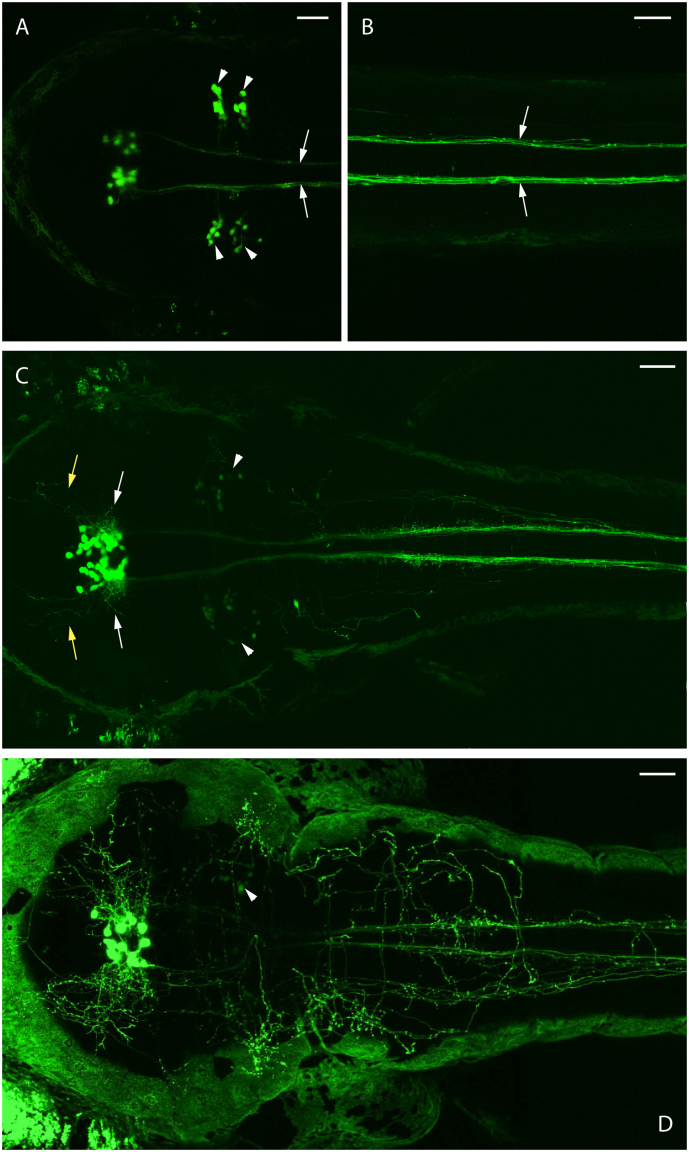
GnRH2 mRNA-expressing neurons could be identified as early as 24 hours post-fertilization (hpf) through whole-mount in situ hybridization (ISH)28. In our transgenic fish, eGFP fluorescence-expressing cells could be observed in the tegmentum as early as 36 hpf. At 2 days post-fertilization (dpf), strong eGFP fluorescence was observed, with the neurons projecting long ipsilateral descending axons (Figures 2A & B, arrows) along the ventral medulla oblongata and into the ventral spinal cord. At 3 dpf, more eGFP positive cells were observed, along with additional lateral- and anterior-extending fibers (Figure 2C). Additional groups of reticulospinal neurons were also identified as GnRH2:eGFP Tg expressing cells in 2 dpf embryos (Figure 2A, arrowheads). However, we failed to detect GnRH2 expression in these reticulospinal neurons by ISH at stages ranging from 2 dpf to 12 dpf. We noticed that the expression level was dramatically decreased in 3 dpf embryos (Figure 2C, arrowheads). In 4 dpf embryos (Figure 2D, arrowhead) noticeably fewer eGFP positive reticulospinal neurons were observed compared with 3 dpf embryos, and these neurons became non-detectable in 8 dpf larvae (data not shown). Therefore, one can reasonably propose the fluorescence signals observed in larvae older than 4 dpf are mostly from tegmentum cells. We did notice that the eGFP brightness of the reticulospinal neurons is significantly dimmer than the tegmentum cells even in 2 dpf embryos (Figure 2A). And the brightness of the eGFP signals does not reflect faithfully their relative expression levels due to the fact that those reticulospinal neurons are located much more superficially, which results in the collection of more fluorescent signals by the confocal microscope. A similar pattern of reticulospinal neuron expression was also reported in GnRH2 transgenic medaka fish29, but it is not clear whether they are sending projections into the spinal cord based on the published data. In zebrafish, we found that most (if not all) of the spinal cord eGFP signals were localized to projections from tegmentum GnRH2 cells (Figures 2A–C) but not the spinal interneurons or reticulospinal neurons as reported in transgenic medaka fish29. We therefore conclude that zebrafish GnRH2 is likely expressed temporarily in the reticulospinal neurons at approximately 2 dpf, and in the remainder of this paper, the tegmentum GnRH2 cells will be simply named “GnRH2 cells”.
Figure 2. Transient expression of GnRH2:eGFP in reticulospinal neurons.
Endogenous eGFP expression in a single GnRH2:eGFP Tg embryo (anterior to the left). The strongest reticulospinal eGFP signals (arrowheads) were observed at 2 dpf (A), and at this stage, long descending spinal cord projections (arrows) from tegmentum GnRH2 cells have been formed (B). The reticulospinal eGFP signals (arrowheads) decreased dramatically at 3 dpf, and additional lateral- (white arrows) and anterior-extending fibers (yellow arrows) were formed (C). The reticulospinal eGFP signals further became barely detectable (arrowhead) at 4 dpf (D). Scale bars = 50 μm.
Architecture of the larval GnRH2 projections
Once the GnRH2 neurons start the dorsal spinal cord projections at 3 dpf, they initially spread out horizontally in the midbrain (Figure 2C, white arrows) and anteriorly into the forebrain (Figure 2C, yellow arrows). Some of the horizontally-spreading projections further curve up along the ventral optical tectum (Figures 3A, asterisk) and form clusters covering the ventral optical tectum (Figure 3B, asterisk). Some further turn posteriorly to form the hindbrain dorsal projections (Supplementay movie 1 and Fig. 3A, boxed areas). Analysis of a 4 dpf live GnRH2:eGFP Tg larva revealed extensive commissural projections in the habenula area (notably crossing the posterior commissure) and bilateral projections proceeding anteriorly through the telencephalon into the olfactory bulbs, forming two clusters of neural fibers that terminate in the anterior ends of the olfactory bulbs (OB, Figure 3A and close-up in 3B at 9 dpf). Dorsal hindbrain projections gradually converge with ventral projections while they are entering the spinal cord. Interestingly, the right side (always dorsal view in this study) projections continue more posterior than the left side projections (Figure 3A, arrowheads). These projections cross the midline in the hindbrain (Figure 3A, yellow arrows) and interact with ventral projections to form extensive networks (Movie S1). In addition, contralateral projections were observed ventrally to the GnRH2 cell bodies (Figures 3C & D, arrow and movie S1) and in the posterior commissure area (Figures 3A & B, white arrows), which is consistent with a previous finding in the European sea bass (Dicentrarchus labrax)30. These observations provide a clear comprehensive description of GnRH2 circuits during zebrafish development.
Figure 3. A stable GnRH2:eGFP Tg line revealing the entire GnRH2 circuit.
All images are dorsal views with anterior (head) to the left. (A) Confocal imaging at 3 dpf showing GnRH2 cell bodies in the midbrain and extensive projections in hindbrain (HB), spinal cord (SC) and telencephalon (Tel), including the habenular commissure and the most anterior parts of the olfactory bulbs (OB). Arrowheads show the ending points of GnRH2 dorsal and ventral projections in the hindbrain and spinal cord. Yellow arrows show the dorsal projections that cross the midline. (B) A close-up confocal image at 9 dpf showing more progressive GnRH2 neuronal projections into the telencephalon and the two olfactory bulbs compared with 5 dpf. The asterisk shows the projections covering the ventral tectum. In A and B, green areas on both sides of the larva correspond to skin or eye autofluorescence. (C) Magnification of Plate B. Commissural projections are observed ventrally to the GnRH2 cell body clusters (arrow) and in the anterior telencephalon (A & B, arrow, commissure anterior). (D) Transient expression of GnRH2:eGFP transgene, allowing the observation of a single GnRH2 neuron harboring both commissural (arrow) and ipsilateral processes. Scale bars = 50 μm.
Complexity of projection patterns
Although the overall projection pattern has been determined, it is still uncertain if all GnRH2 cells are sending projections into all the target locations as described above. We extended this study by labeling individual GnRH2 neurons using transient expression of the GnRH2:eGFP construct and subsequently observed high individual neuron target variability. Ipsilateral primary projections extended posteriorly up to ~600 μm beyond the end of the yolk sac by 36 hpf (Figures 4A & B). In addition, highly branched axon collaterals could be observed at 4 dpf, and very thin primary collaterals were observed to branch perpendicularly off the stem axon at regular intervals along the spinal cord (Figure 4C, arrows). This has not been revealed in the transgenic larvae possibly because multiple projections cluster to form bundles that can easily mask the details of individual projections. Most notably, hindbrain dorsal projections could not be observed for some GnRH2 neurons (Figure 4C). On the other hand, some GnRH2 neurons showed no ventral spinal cord projections. Instead, they send out only the dorsal hindbrain projections (Figures 4D & E), some of which (if not all) will eventually enter the spinal cord (data not shown). Furthermore, we observed that, in a few 4 dpf larvae, most neurons do not form nerve terminals in the olfactory bulb (Figure 4E), and the nerve terminals always project from the GnRH2 cells on the same side (projecting ipsilaterally), left side from the left side GnRH2 cell (Figure 4C, arrowhead) and right side from right side GnRH2 cell (Figure 4D, arrowhead). We obtained about 40 single neuron-labeled larvae and observed only the above four described patterns. Since each larva has about 30–40 labeled neurons, we have reason to believe that there are other projection patterns of individual GnRH2 neurons that have not been observed here. From this study, at least, different combinations of target choices by processes arising from individual GnRH2 neurons were identified.
Figure 4. Single cell-labeling of GnRH2:eGFP neurons.
GnRH2:eGFP axon growth reaches ~1000 μm by 36 hpf (A & B). At 4 dpf, some larvae with left side-labeled single GnRH2 neurons show clusters of left side telencephalon fibers (arrowhead) and commissural projections (arrows) running across the midline in the spinal cord (C). Other larvae displayed left side-labeled single GnRH2 neurons showing clusters of left side telencephalon fibers (arrowhead) and dorsal hindbrain projections but not spinal cord projections (D). Larvae with similar projections as shown in (E) that do not project into the olfactory bulb. Scale bars = 50 μm.
Architecture of the juvenile GnRH2 projections
In 22 dpf GnRH2:eGFP Tg juveniles (Figures 5A–H), consistent with what we observed at the early developmental stages (Figures 2C and 3A & B and movie S1), GnRH2 cells are located only in the midbrain tegmentum, send out mainly lateral projections (Figures 5A & B, arrows), and further extend to the bottom of the tectum covering the white matter (Figures 5A & B, arrowheads). Projections can be easily classified into dorsal (Figures 5C–H, white arrows) and ventral (Figures 5C–G, yellow arrows) groups. The right side dorsal projections consistently end sooner than the left side projections in the spinal cord (Figure 5G), and they also converge with ventral projections sooner than the left side dorsal projections at larval stages (Figure 3A, arrowheads), suggesting that the dorsal projections that converge sooner with ventral projections also end sooner in the spinal cord. Both side secondary projections eventually end in the posterior spinal cord (Figure 5H). This study indicates that the overall GnRH2 projection pattern has been well established at approximately 4 dpf.
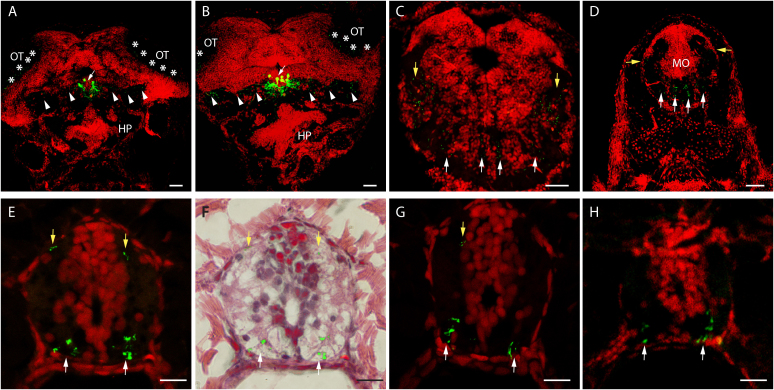
Figure 5. Juvenile GnRH2:eGFP Tg neurons.
Confocal imaging of 7 μm transversal sections from a stable 22 dpf GnRH2:eGFP Tg fish (reconstructed stacks of 0.5 or 1 μm sections). Images are presented in the order of anterior to posterior (A to H). Nuclei are stained red via PI staining. (F) A hematoxylin and eosin staining of section E, overlapped with the staining from section E following the removal of the red channel. Scale bars: 100 μm (A–C) and 10 μm (D–H). HP, hyphothalamus; OT, optical tectum; MO, medulla oblongata.
Simultaneous evaluation of the GnRH2 and GnRH3 systems in double transgenic larval and juvenile zebrafish
Different GnRH forms have been reported to trigger similar events with different efficiencies17,19,23,31. In order to investigate potential interactions of the GnRH2 neurons with the hypophysiotrophic isoform, GnRH3, in zebrafish, we believe it is necessary to delineate both the GnRH2 and GnRH3 neural systems simultaneously. Consequently, we crossed our GnRH2:eGFP Tg line with a stable GnRH3:tdTomato Tg transgenic line developed in our lab and obtained a stable, double transgenic zebrafish line expressing both eGFP (green) and tdTomato (red) in putative GnRH2 and GnRH3 neurons, respectively. This enabled us to track both GnRH2 and GnRH3 soma and projections simultaneously in live zebrafish embryos and larvae.
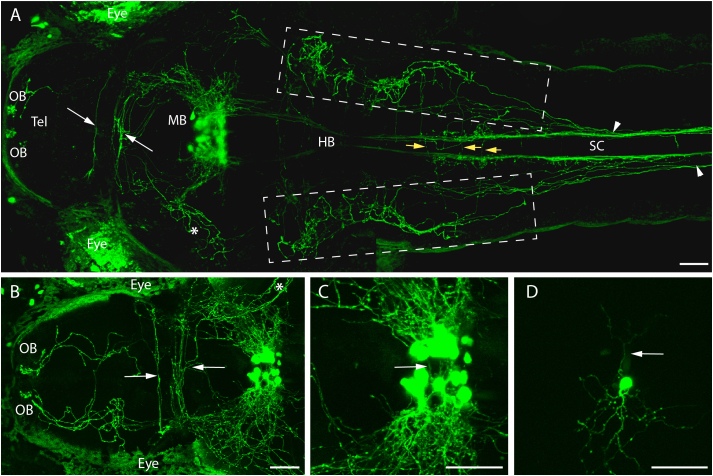
Similarly to GnRH2 neurons (Figures 3A & B), by 4 dpf, GnRH3 neurons project extensive fibers throughout the brain, and the projections cross the posterior commissure (Figure 6A, arrowheads). After dissecting the brain out of the larva, GnRH2 nerve terminals can be clearly localized to the posterior olfactory areas (Figure 6B). A clear difference between the two GnRH systems was observed in the midbrain tectum (Figure 6C, 35 degrees away from dorsal axis; also see 3D view in movie S2), where GnRH3 projections mainly form two layers of fiber clusters, one at roughly the same level as the GnRH2 layer (Figure 6C, arrowhead) and the other one on the top of the tectum (Figure 6C, arrow). The GnRH3 projections form a cluster of neural networks that enclose the tectum, as opposed to the more ventral GnRH2 projections (Figures 6A & C). Contrary to GnRH2 neurons, we observed that GnRH3 neurons have more abundant dorsal projections than ventral projections in the midbrain, hindbrain and spinal cord (Figures 6A, C–F). Similarly to GnRH2, the dorsal right hindbrain GnRH3 projections appear to converge with the ventral projections earlier than the left dorsal side while entering the spinal cord (Figure 6D). This architecture of GnRH projections is further confirmed by observations made in 22 dpf juvenile transgenic zebrafish (Figures 7A–H, anterior to posterior), except that additional groups of GnRH3:tdTomato expressing neurons were identified in the hindbrain (Figure 7B, arrows). No potential GnRH2 afferent connections were observed from GnRH3 fibers (Figure 6A). The relative abundance of GnRH3 dorsal projections in the larval spinal cord (Figure 6F) is sustained in juvenile zebrafish (Figures 7C–G), and the dorsal projections extend further in the spinal cord than the ventral projections. Similarly to GnRH2 projections, the side with the abundant projections continues more posteriorly than the side with less abundant projections. The dorsal right GnRH3 projections, which converge with ventral projections sooner than the dorsal left side (Figure 6D), also terminate sooner than the left side (Figures 7 D–F). All GnRH3 projections, except the dorsal left, end sooner than the same side GnRH2 projections (Figure 7E). The dorsal left GnRH3 projections extend further posteriorly in the spinal cord than both sides of GnRH2 dorsal projections (Figures 7F–G) but not as long as the GnRH2 ventral projections (Figure 7H), which extend ~800 μm longer than the longest GnRH3 projections (Figure 7G & H, quantified by counting the number of continuous sections in between).
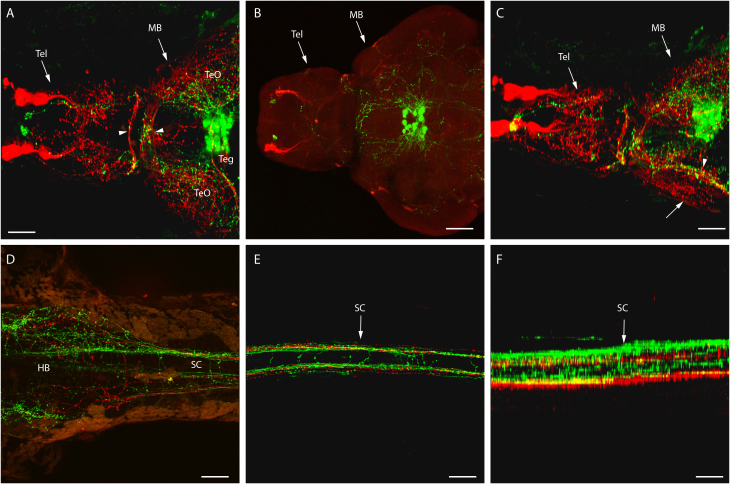
Figure 6. A stable 4 dpf double GnRH2:eGFP and GnRH3:tdTomato Tg line reveals the entire GnRH circuit of a living larva.
Double transgenic showing both GnRH2 (green) and GnRH3 (red) circuits at 4 dpf, using confocal microscopy (anterior to the right, unless specified otherwise). (A) Dorsal view of both GnRH2 and GnRH3 cell bodies and projections in midbrain including tegmentum (Teg) and tectum opticums (TeO), posterior commissure area (arrowheads), and telencephalon (Tel). (B) A dorsal view of a dissected double transgenic brain, and (C) a slight right view (35 degree from the dorsal axis) of (A) showing the relative positions of GnRH3 projections (arrow) with GnRH2 projections (arrowhead). Dorsal view of both GnRH2 and GnRH3 projections in hindbrain (D) and spinal cord (E). Image (F) provides a right-side lateral rendering of (E). Tel, telencephalon; MB, midbrain; HB, hindbrain. SC, spinal cord. Scale bars = 50 μm.
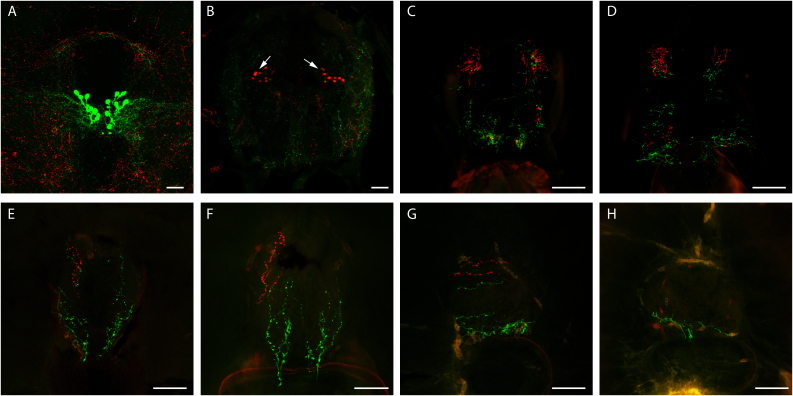
Figure 7. Projections of both GnRH2:eGFP and GnRH3:tdTomato neurons in juvenile zebrafish.
Confocal imaging of 100 μm transversal sections from a 22 dpf stable double GnRH2:eGFP (green) and GnRH3:tdTomato (red) Tg fish (reconstructed stacks of 2 to 4 μm sections). Images A through H are from the anterior (the tegmentum) to the posterior region of the trunk. Scale bars = 50 μm.
Architecture of the adult GnRH2 projections
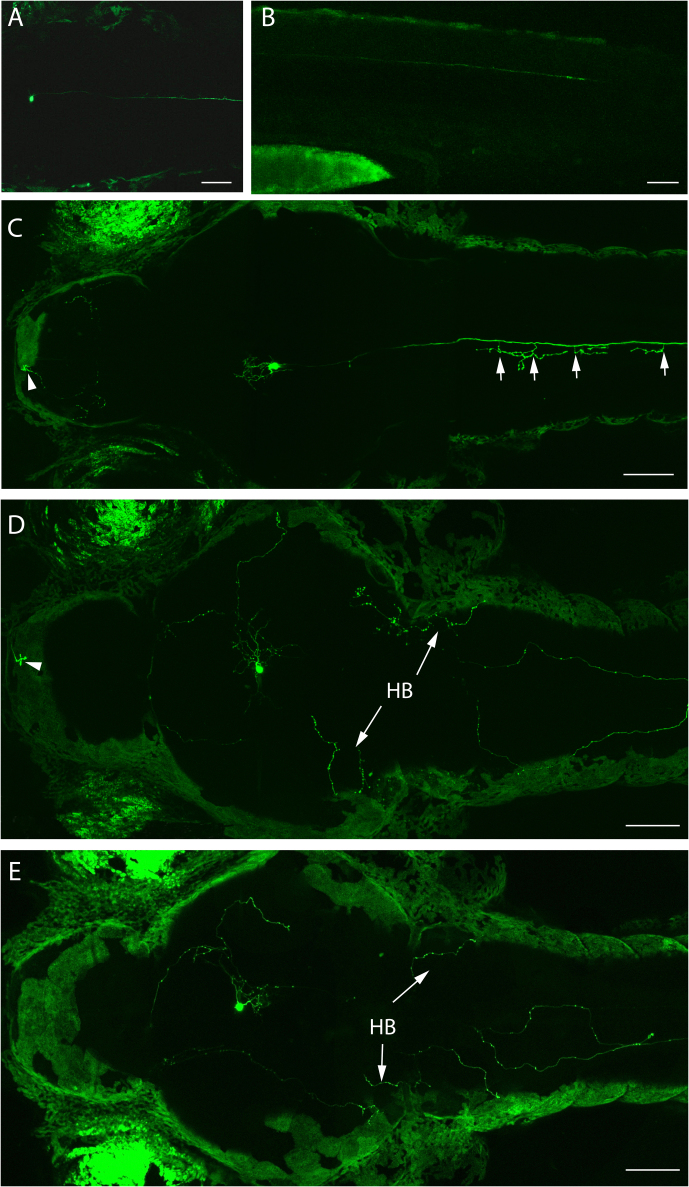
Although a well-established pattern of GnRH2 fibers is formed at very early stages, due to the possible changes of GnRH2 projection patterns during development and adulthood32, we thought it was necessary to map the GnRH2 projections in adult fish. In the brain of adult GnRH2:eGFP Tg fish, consistent with what we observed in the larvae, sagittal sections revealed that the most anterior GnRH2 projections end in the olfactory bulbs (Figure 8A), and no fluorescent signals could be detected in the olfactory epithelium (Figure 8A), as opposed to GnRH3 neurons (Figure S1A & B). The projections in the olfactory bulbs are widely distributed, only with more intensive projections in the area that is proximal to the midline (Figure 8B, enclosed by dashed box). More confined GnRH2 projections were found in the telencephalon, corresponding to the middle and ventral telencephalon, and are also close to the subpallium midline (Figure 8C, enclosed area). As expected, the most prominent projections were detected in the midbrain tegmentum (Figure 8D), and the cell bodies are organized as two compact clusters facing each other with very little (if any) contralateral projections between clusters. GnRH2 fibers were also observed around the tectal ventricule in the periventricular gray zone of the optic tectum with less stratum marginale projections in the posterior tectum (Figures 8D & E). In the medulla oblongata, the projections are also widely distributed (Figure 8F).
Figure 8. Adult zebrafish GnRH2 circuit.
(A) Sagittal section (anterior to the left) of olfactory epithelium and olfactory bulb area. (B–F) Confocal imaging of 100 μm cross-brain sections from a stable GnRH2:eGFP Tg adult fish (reconstructed stacks of 2 to 4 μm sections), showing the GnRH2 projections in the olfactory bulb (B), telencephalon (C), mid-diencephalon (D), posterior diencephalon (E) and medulla oblongata (F). OE, olfactory epithelium; OB, olfactory bulb; SN, solitary nucleus; HP, hypothalamus. Dashed boxes show the areas with the most intensive projections. Scale bars = 100 μm.
GnRH2 neurons send projections into the pituitary
We noticed much more intensive and directional GnRH2 projections in the mediobasal hypothalamus (notably the nucleus of the ventral hypothalamus), most likely pointing to the pituitary stalk (Figure 8D, arrows versus Figure 8E). This was confirmed by a sagittal section of this area from a double GnRH3:tdTomato and GnRH2:eGFP Tg fish (Figure S3C, arrow), which led us to consider the possibility of GnRH2 projections into the pituitary. One clear anatomical difference between the GnRH3 and GnRH2 neurons is considered to be that the former directly projects to the pituitary, and the latter projects widely throughout the brain but not to the pituitary33,34. Our lab previously reported that the GnRH2 peptide exists in pooled zebrafish pituitary extracts, as detected by ELISA analysis, albeit at a much lower concentration compared with GnRH335. Our newly established GnRH2:eGFP Tg fish provided us the opportunity to readdress this question in a more straightforward manner. Clearly visible projections were observed in transgenic pituitary sections (Figure 9A) using a confocal microscope, but the intensity is much lower than GnRH2 neurons in other parts of the brain. Since over-expression of GnRH2 has been shown to suppress food intake and excessive feeding could increase GnRH2 expression in the hyphothalamus32,36, we considered that food abundance might also affect the GnRH2 projection pattern. Therefore, adult fish were starved for 14 days, followed by brain collections to observe the GnRH2 neuronal projections. As shown in Figures 9B–D, a dramatic increase of projections was observed in the pituitary (but not in other brain parts; data not shown) of fasted fish. Together, these data indicate that GnRH2's potential role of regulating zebrafish feeding behavior might also involve the participation of the pituitary.
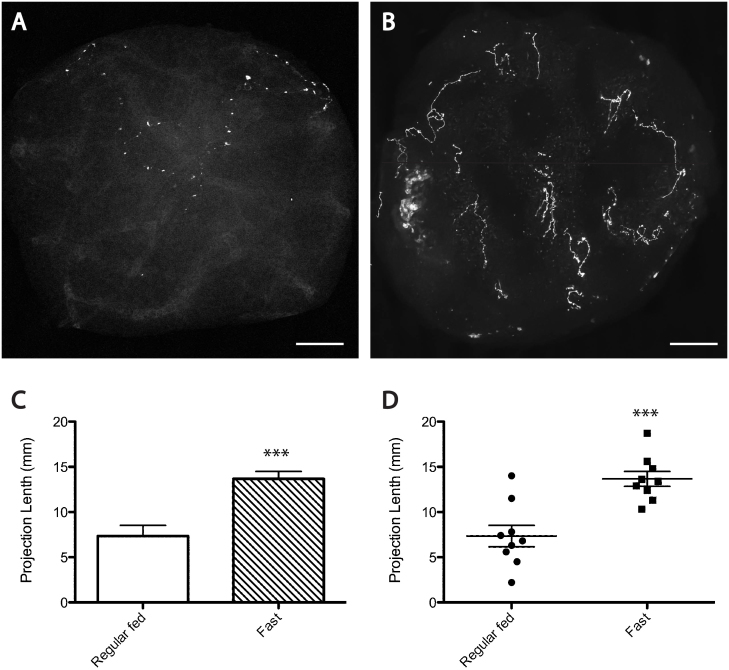
Figure 9. Fasting of adult zebrafish resulted in significantly more projections of GnRH2 neurons in the pituitary.
Compared with the projections in control fish pituitaries (A), the fasted zebrafish pituitary showed dramatically more eGFP-positive fibers at a similar position (B). The projecting lengths of series of 100 μm sections were added together for each pituitary in both control and fasted groups, and a bar graph (C) and scatter plot (D) were designed to give the overall difference of the projection lengths in both experimental groups. Data represent mean ± SEM. Statistical significance was evaluated using a two-tailed unpaired student's t test. ***P < 0.0005%. Scale bars = 50 μm.
Discussion
Most of the currently proposed functions of GnRH2 are concerned directly or indirectly with reproduction37,38,39,40, despite its wide distribution outside the reproductive system. The exact function of the GnRH2 system remains largely elusive, due, at least partially, to the lack of GnRH2 gene in the mouse model, where powerful genetic approaches, especially gene knockout techniques, are available for functional genetic studies. There is a strong need for a proper model organism for investigating GnRH2 functions. Zebrafish has the same two-gene GnRH system as primates, and with its translucent embryos and larvae, simple central nervous system, rapid development and multiple available genetic tools (comparable to mice), it is an excellent model for GnRH2 functional studies. Existing studies suggest that GnRH2 regulates animal food intake and sexual behavior40,41,42,43, and we believe that understanding the anatomy, localization, ontogeny and projection pattern of the GnRH2 neurons is essential to evaluate their modulatory input into circuits that control these behaviors. By using BAC DNA that contains long regulative elements and coupling it with Tol2 technology, we obtained, for the first time, a stable GnRH2:eGFP Tg line. The co-staining with both ISH (Figure S1) and ICC (Figures 1D–E2) indicates that this line specifically labels the GnRH2 neurons and fibers with eGFP protein. Although ICC can also stain the cells and fibers, there are some inherent disadvantages of the immunostaining procedure: (i) it has a poor signal-noise ratio compared to endogenous eGFP signals27 and (ii) it lacks the benefit of live embryo imaging44. Taken together, the advantages of the GnRH2:eGFP Tg line (i.e., better S/N and live imaging benefits) make it more suitable for observing the GnRH2 neuronal projections. Using this line, we generated the first complete view of the projection patterns that originate from GnRH2 cells in a vertebrate from early developmental stages to adulthood.
First, based on the GnRH2:eGFP Tg line, both GnRH2 transcription and the GnRH2 cell behaviors could be easily monitored in real time without sacrificing the fish, and the highly sensitive eGFP reporter enabled us to confirm the major site of GnRH soma in the midbrain tegmentum and revealed previously undescribed groups of reticulospinal cells in the hindbrain. The single cell analysis of GnRH2 cells also revealed the complexity of the GnRH2 cell groups, and individual neurons exhibited high variability in their projection pathways and targets. The observed diversity may represent true additional genetically defined subtypes of GnRH2 neurons that may be associated with different physiological functions. At 4 dpf, most fluorescent signals are from the tegmentum cells, which could further be classified into at least two groups: the ventral spinal cord-projecting cells and the dorsal spinal cord-projecting cells, based on the fact that we did not detect any single eGFP-labeled neurons distributing both dorsal and ventral spinal cord projections (Figures 4C–E). Since more eGFP-expressing cells were observed in 3 and 4 dpf larvae compared with 2 dpf larvae (Figures 2A & D), the ventral spinal cord-projecting cells seem to form earlier than the dorsal spinal cord-projecting cells. The putative proximity to the spinal motor neurons in the ventral spinal cord of those ventral spinal cord-projecting GnRH2 cells (Figures 5E–H) suggests that these are descending motor neurons, which send out one of the earliest formed axon tracts in the developing central nervous system45. The GnRH2 neurons wide dendritic projections in the central nervous system indicate that they might be interacting with other spinal cord-projecting neurons to ensure the processing of sensory information in the brain and transmission to the spinal cord, ultimately initiating the appropriate behavioral motor output by sending signals to the motor neurons. This could be a conserved characteristic across species, and the conserved GnRH2 gene might play important roles in regulating animal behaviors. Further behavioral and physiological analyses comparing GnRH2-depleted and wild-type control larvae may help to test this hypothesis.
Our comprehensive analysis of GnRH2:eGFP expression patterns at different developmental stages revealed a very complex projection pattern of GnRH2 neurons. We developed, for the first time, a double transgenic zebrafish with both GnRH2 and GnRH3 neurons labeled with eGFP and tdTomato, respectively. Using this tool, we were able to observe both GnRH systems in vivo simultaneously in the same animal. Moreover, we found that very well-established axon distributions can be observed as early as 4 dpf for both GnRH neurons. At this stage, GnRH2 fibers are located widely throughout the central nervous system (CNS), initiating from the tegmentum and extending posteriorly as far as the caudal spinal cord and anteriorly as far as the olfactory bulbs. GnRH2 fibers end at the olfactory nerve without entering the olfactory epithelium, while GnRH3 was found to send fibers into olfactory bulbs through the olfactory nerve system. These data suggest that, if GnRH2 and GnRH3 are involved in detecting and transducing environmental odor cues46, which still needs additional data, GnRH2 is not supposed to be directly responsible for detecting odors. The GnRH2 spinal cord projections can be divided as dorsal and ventral groups anatomically. We found that GnRH2 dorsal secondary projections do not extend as far as ventral projections, which extend almost the entire length of the spinal cord. Since most motor neurons are located ventrally and most sensory neurons are located dorsally, our results suggest that GnRH2 might be involved in both afferent (input) and efferent (output) signal processing in the central nervous system. Additionally, they are very likely to be performed by different GnRH2 cell groups. Compared with GnRH3 neuronal projections, which are predominantly dorsally directed, we propose that GnRH2 neurons may be more involved in conveying motor information away from the CNS to the spinal cord, and GnRH3 neurons may be more involved in receiving sensory information from the spinal cord.
Interestingly, we observed distinct projections in the pituitaries of the GnRH2:eGFP Tg line, where GnRH2 projections were believed to not exist. GnRH3 cells send projections into the pituitary to regulate the synthesis and secretion of the gonadotropins through binding to its specific receptors on the gonadotrophs. All known (four) GnRH receptors are reported to be expressed in the zebrafish pituitary19, but their precise distribution and functions remain vague and in need of clarification. The existence of GnRH2 axons in the pituitary indicates that GnRH2 may have important roles in this gland that are distinct from GnRH3 functions through binding to its own putative receptor(s) and/or regulating reproduction-related cells or other pituitary cells. This possibility is further strengthened by the experiment in which the adult zebrafish were fasted. The only obvious difference was observed in the pituitary, where more GnRH2 fibers clearly existed in pituitaries of fasted fish compared with control fish. The hypothalamic-pituitary-adrenal axis has been demonstrated to respond to changes of food intake41, and the pituitary can directly regulate metabolism (converting food into energy) through feeding behavior, physical activity, and energy expenditure42. These findings, together with our results, indicate that GnRH2 may be involved in regulating nutrition-related processes and behaviors through its projections to the pituitary gland. It is equally possible that more pituitary projections are simply a result of the metabolic changes but not a cause. A GnRH2 knockout zebrafish using the popular transcription activator-like effector nuclease (TALEN) technology, coupled with food consumption behavior analysis and molecular analysis of potentially affected genes in the pituitary, may help answer this question.
In conclusion, the establishment of the GnRH2:eGFP Tg zebrafish line has generated a large quantity of data, including novel findings regarding GnRH2 fiber development, the role of GnRH2 as a potential factor important for pituitary functions, and distinct projection patterns that differ from the GnRH3 system. Considering the early and progressive projections into the ventral (motor) roots compared with GnRH3 neurons, some GnRH2 neurons may function as a modulator of energy balance physically by directly controlling the overall motility through its spinal cord projections and indirectly by regulating the metabolism through its projections in the pituitary. The rest of the projections scattered throughout the CNS may act as a sensor to trigger its modulating functions. We believe that our transgenic lines, along with the powerful genetic tools of the zebrafish model, will greatly facilitate future work to elucidate the function(s) of the GnRH2 gene and its potential interactions with GnRH3 at the molecular, cellular and physiological levels.
Methods
Zebrafish maintenance
The spawning of adults and husbandry of embryos were carried out according to standard protocols47. Briefly, 1-phenyl-2-thiourea (PTU) was added to the water every second day during 2–7 dpf at a concentration of 0.2 mM to prevent pigmentation. Larvae used for experiments were anaesthetized with tricaine (MS-222) and either observed with standard/confocal microscopy or fixed in 4% paraformaldehyde for further treatment. Juveniles and adult zebrafish were raised at 28°C on a 12 hour light: 12 hour dark cycle and maintained in a recirculated water system. All procedures were approved by the Institutional Animal Care and Use Committee at the University of Maryland School of Medicine.
Plasmids construction and generation of transgenic zebrafish lines
A DKEYP-87D8 BAC clone containing complete GnRH2 gene and large upstream sequences was ordered from Source BioScience (www.lifesciences.sourcebioscience.com), and BAC modifications were generated according to standard protocols using galK-mediated recombination in bacteria48,49. GnRH2:eGFP was generated by replacing the GnRH2 coding sequence (from start codon to stop codon) with the eGFP coding sequence and adding of a Tol2 cassette.
BAC DNA was injected into one-cell stage wild type (WT) embryos at 50 ng/μl along with 25 ng/μl Tol2 transposase mRNA according to established procedures47. The GnRH2:eGFP was injected into one-cell-stage embryos. Embryos with efficiently labeled GnRH2 neurons were grown up for generating stable F1 transgenic zebrafish. F1 zebrafish were then crossed with WT to make stable F2 transgenics. All GnRH2:eGFP Tg fish used in the following content are F2 transgenic fish, which were crossed with GnRH3:eGFP Tg fish to make double transgenic zebrafish.
Antibody preparation, in situ hybridization, and immunocytochemistry
Anti-GnRH antibodies have been traditionally made against the mature decapeptides or the preproteins, which usually resulted in non-specific binding to multiple GnRH isoforms50,51. GnRH-associated peptides (GAP) have successfully been used to make antibodies that are specific to distinct GnRH isoforms30,52. The cDNA sequence of the zebrafish GnRH2-GAP was obtained by PCR with the following primers:
GAPF: 5′-AAAGAGACATATGGAGATAGACCTCTACGA-3′
GAPR: 5′-GGTGTCACTCGAGCTACTTTCTCTTTTGGAAAT-3′
Using the restriction endonucleases NdeI and XhoI, the sequences were cloned into the pET-15b expression vector and transformed into Rosetta-gami B(DE3)pLysS E. coli cells. Protein expression was induced with 1 mM IPTG, and after dissolving in 8 M urea, the N-terminus His-tagged recombinant protein was purified with NI-NTA columns (Promega) and desalted with G-15 desalting columns (Pharmacia; Stockholm, Sweden). The purified peptides were used to develop GnRH2 polyclonal antibodies in rabbit (Proteintech; Chicago, IL).
Double labeling of ISH and ICC were carried out as described47,53, without proteinase K treatment, using DIG-labeled antisense GnRH2 riboprobes28 previously used by our lab. Mouse anti-eGFP (diluted 1:100; Cell Signaling) was added with anti-DIG POD antibody (diluted 1:500; Roche Diagnostics, Germany) after blocking. After 5 × 10 minute washes in PBS, Fluorescein-conjugated goat anti-mouse (diluted 1:100; Thermo Scientific, USA) was added to detect the anti-eGFP antibody, followed by 3 × 10 minute washes with PBS and then a TSA Plus Cy3 solution treatment. Rabbit GnRH2 antibody (1:500) was used on fixed GnRH2:eGFP Tg fish brain sections. Cy3-labeled goat anti-rabbit antibody (1:200, KPL) was added to stain GnRH2 protein-expressing cells with red fluorescence. VECTASHIELD HardSet Mounting Medium (Vector Laboratories, USA) was used to mount the slides.
Antibody pre-absorption was performed as described previously54. Briefly, 0.2% GnRH2 antibody was incubated with ~10 μg/ml purified overexpressed GnRH2-GAP fusion protein at 4°C for 16 h in 0.5% non-fat milk powder/PBST solution with mild agitation. After centrifuging the antigen-antibody mixture at 16,000 × g for 20 min, the supernatant was carefully pipetted into a clean vial and used in the immunocytochemistry analysis.
Fluorescent microscopy
Fluorescence expression was initially assayed using a Zeiss Axioplan 2 microscope (Carl Zeiss MicroImaging, Inc., Thornwood, NY, USA). For more detailed analysis of fluorescent protein expression and projections, larvae were mounted in 1.0% low-melt agarose with tricaine and imaged with a Zeiss Radiance 2100 laser scanning system together with Laser-Sharp and LSM imaging software (Carl Zeiss MicroImaging, Inc.). Images were processed using Adobe Photoshop CS2. The extensive GnRH neural projections were, at times, obscured by skin or eye autofluorescence when viewed as confocal projections. Therefore, for Figures 5 and 6, we used ImageJ (http://rsb.info.nih.gov/ij; developed by Wayne Rasband, NIH) to manually edit each z-slice and remove autofluorescence and background fluorescence, taking care always to spare nearby axons. Movies were made with Fluorender software (http://www.sci.utah.edu/software/46-documentation/137-fluorender.html). Reconstructed three-dimensional images were projected to a standardized view for figure presentation. Projection lengths were measured using ImageJ on each 100 μm section and were added together for each individual pituitary.
Fasting of adult zebrafish and sampling
Individual adult fish were kept in separate tanks in which water was replaced every day for 14 days. The control groups were fed normally with commercial fish flakes, and the fasted groups were not given any food. After 14 days, all fish were anesthetized with MS-222, and the brain and pituitary were dissected and fixed in 4% paraformaldehyde at 4°C overnight. Fluorescence was observed using confocal microscopy directly after sectioning.
Author Contributions
W.X. performed most experiments, O.S. and N.Z. generated the GnRH2-GAP antibody, and S.X. performed the antibody pre-absorption analysis. W.X. and Y.Z. wrote the manuscript and all authors discussed the results and commented on the manuscript.
Supplementary Material
Supplementary Information
Supplementary Video 1
Supplementary Video 2
Acknowledgments
We thank John Stubblefield for critical reading and English proofing, and Dr. Tian-E Chen from Johns Hopkins for helping with confocal imaging.
References
- Baba Y., Matsuo H. & Schally A. V. Structure of the porcine LH- and FSH-releasing hormone. II. Confirmation of the proposed structure by conventional sequential analyses. Biochem Biophys Res Commun 44, 459–63 (1971). [DOI] [PubMed] [Google Scholar]
- King J. A. & Millar R. P. Comparative aspects of luteinizing hormone-releasing hormone structure and function in vertebrate phylogeny. Endocrinology 106, 707–17 (1980). [DOI] [PubMed] [Google Scholar]
- Iwakoshi E. et al. Isolation and characterization of a GnRH-like peptide from Octopus vulgaris. Biochem Biophys Res Commun 291, 1187–93 (2002). [DOI] [PubMed] [Google Scholar]
- Adams B. A. et al. Six novel gonadotropin-releasing hormones are encoded as triplets on each of two genes in the protochordate, Ciona intestinalis. Endocrinology 144, 1907–19 (2003). [DOI] [PubMed] [Google Scholar]
- Powell J. F. et al. Two new forms of gonadotropin-releasing hormone in a protochordate and the evolutionary implications. Proc Natl Acad Sci U S A 93, 10461–4 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tostivint H. Evolution of the gonadotropin-releasing hormone (GnRH) gene family in relation to vertebrate tetraploidizations. Gen Comp Endocrinol 170, 575–81 (2011). [DOI] [PubMed] [Google Scholar]
- Zohar Y., Munoz-Cueto J. A., Elizur A. & Kah O. Neuroendocrinology of reproduction in teleost fish. Gen Comp Endocrinol 165, 438–55 (2010). [DOI] [PubMed] [Google Scholar]
- Okubo K. & Nagahama Y. Structural and functional evolution of gonadotropin-releasing hormone in vertebrates. Acta Physiol (Oxf) 193, 3–15 (2008). [DOI] [PubMed] [Google Scholar]
- Muske L. E. Evolution of gonadotropin-releasing hormone (GnRH) neuronal systems. Brain Behav Evol 42, 215–30 (1993). [DOI] [PubMed] [Google Scholar]
- Roch G. J., Busby E. R. & Sherwood N. M. Evolution of GnRH: diving deeper. Gen Comp Endocrinol 171, 1–16 (2011). [DOI] [PubMed] [Google Scholar]
- Stewart A. J., Katz A. A., Millar R. P. & Morgan K. Retention and silencing of prepro-GnRH-II and type II GnRH receptor genes in mammals. Neuroendocrinology 90, 416–32 (2009). [DOI] [PubMed] [Google Scholar]
- White R. B., Eisen J. A., Kasten T. L. & Fernald R. D. Second gene for gonadotropin-releasing hormone in humans. Proc Natl Acad Sci U S A 95, 305–9 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong T. T. et al. Developmental expression of three forms of gonadotropin-releasing hormone and ontogeny of the hypothalamic-pituitary-gonadal axis in gilthead seabream (Sparus aurata). Biol Reprod 71, 1026–35 (2004). [DOI] [PubMed] [Google Scholar]
- Soga T., Ogawa S., Millar R. P., Sakuma Y. & Parhar I. S. Localization of the three GnRH types and GnRH receptors in the brain of a cichlid fish: Insights into their neuroendocrine and neuromodulator functions. J Comp Neurol 487, 28–41 (2005). [DOI] [PubMed] [Google Scholar]
- Ikemoto T. & Park M. K. Molecular and evolutionary characterization of the GnRH-II gene in the chicken: distinctive genomic organization, expression pattern, and precursor sequence. Gene 368, 28–36 (2006). [DOI] [PubMed] [Google Scholar]
- Hu S. Y. et al. Cloning and functional analysis of the proximal promoter region of the three GnRH genes from the silver sea bream (Sparus sarba). Comp Biochem Physiol B Biochem Mol Biol 151, 373–80 (2008). [DOI] [PubMed] [Google Scholar]
- Chen A. et al. Two forms of gonadotropin-releasing hormone (GnRH) are expressed in human breast tissue and overexpressed in breast cancer: a putative mechanism for the antiproliferative effect of GnRH by down-regulation of acidic ribosomal phosphoproteins P1 and P2. Cancer Res 62, 1036–44 (2002). [PubMed] [Google Scholar]
- Parker J. D., Malik M. & Catherino W. H. Human myometrium and leiomyomas express gonadotropin-releasing hormone 2 and gonadotropin-releasing hormone 2 receptor. Fertil Steril 88, 39–46 (2007). [DOI] [PubMed] [Google Scholar]
- Tello J. A., Wu S., Rivier J. E. & Sherwood N. M. Four functional GnRH receptors in zebrafish: analysis of structure, signaling, synteny and phylogeny. Integr Comp Biol 48, 570–87 (2008). [DOI] [PubMed] [Google Scholar]
- Morimoto C. et al. GnRH II as a possible cytostatic regulator in the development of endometriosis. Hum Reprod 20, 3212–8 (2005). [DOI] [PubMed] [Google Scholar]
- Serin I. S. et al. GnRH-II mRNA expression in tumor tissue and peripheral blood mononuclear cells (PBMCs) in patients with malignant and benign ovarian tumors. Eur J Obstet Gynecol Reprod Biol 149, 92–6 (2010). [DOI] [PubMed] [Google Scholar]
- Lin Y. M. et al. Transcripts of testicular gonadotropin-releasing hormone, steroidogenic enzymes, and intratesticular testosterone levels in infertile men. Fertil Steril 90, 1761–8 (2008). [DOI] [PubMed] [Google Scholar]
- Islami D., Chardonnens D., Campana A. & Bischof P. Comparison of the effects of GnRH-I and GnRH-II on HCG synthesis and secretion by first trimester trophoblast. Mol Hum Reprod 7, 3–9 (2001). [DOI] [PubMed] [Google Scholar]
- Cheon K. W., Lee H. S., Parhar I. S. & Kang I. S. Expression of the second isoform of gonadotrophin-releasing hormone (GnRH-II) in human endometrium throughout the menstrual cycle. Mol Hum Reprod 7, 447–52 (2001). [DOI] [PubMed] [Google Scholar]
- Perfito N., Zann R., Ubuka T., Bentley G. & Hau M. Potential roles for GNIH and GNRH-II in reproductive axis regulation of an opportunistically breeding songbird. Gen Comp Endocrinol 173, 20–6 (2011). [DOI] [PubMed] [Google Scholar]
- Budick S. A. & O'Malley D. M. Locomotor repertoire of the larval zebrafish: swimming, turning and prey capture. J Exp Biol 203, 2565–79 (2000). [DOI] [PubMed] [Google Scholar]
- Kaddoum L., Magdeleine E., Waldo G. S., Joly E. & Cabantous S. One-step split GFP staining for sensitive protein detection and localization in mammalian cells. Biotechniques 49, 727–8, 730,, 732 passim (2010). [DOI] [PubMed] [Google Scholar]
- Palevitch O. et al. Ontogeny of the GnRH systems in zebrafish brain: in situ hybridization and promoter-reporter expression analyses in intact animals. Cell Tissue Res 327, 313–22 (2007). [DOI] [PubMed] [Google Scholar]
- Kusakabe T. G. et al. A conserved non-reproductive GnRH system in chordates. PLoS One 7, e41955 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servili A. et al. The Highly conserved gonadotropin-releasing hormone-2 form acts as a melatonin-releasing factor in the pineal of a teleost fish, the european sea bass Dicentrarchus labrax. Endocrinology 151, 2265–75 (2010). [DOI] [PubMed] [Google Scholar]
- Millar R. P. GnRH II and type II GnRH receptors. Trends Endocrinol Metab 14, 35–43 (2003). [DOI] [PubMed] [Google Scholar]
- Nishiguchi R., Azuma M., Yokobori E., Uchiyama M. & Matsuda K. Gonadotropin-releasing hormone 2 suppresses food intake in the zebrafish, Danio rerio. Front Endocrinol (Lausanne) 3, 122 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda S. et al. Regular pacemaker activity characterizes gonadotropin-releasing hormone 2 neurons recorded from green fluorescent protein-transgenic medaka. Endocrinology 151, 695–701 (2010). [DOI] [PubMed] [Google Scholar]
- Gonzalez-Martinez D. et al. Immunohistochemical localization of three different prepro-GnRHs in the brain and pituitary of the European sea bass (Dicentrarchus labrax) using antibodies to the corresponding GnRH-associated peptides. J Comp Neurol 446, 95–113 (2002). [DOI] [PubMed] [Google Scholar]
- Steven C. et al. Molecular characterization of the GnRH system in zebrafish (Danio rerio): cloning of chicken GnRH-II, adult brain expression patterns and pituitary content of salmon GnRH and chicken GnRH-II. Gen Comp Endocrinol 133, 27–37 (2003). [DOI] [PubMed] [Google Scholar]
- Kauffman A. S., Bojkowska K., Wills A. & Rissman E. F. Gonadotropin-releasing hormone-II messenger ribonucleic acid and protein content in the mammalian brain are modulated by food intake. Endocrinology 147, 5069–77 (2006). [DOI] [PubMed] [Google Scholar]
- Maney D. L., Richardson R. D. & Wingfield J. C. Central administration of chicken gonadotropin-releasing hormone-II enhances courtship behavior in a female sparrow. Horm Behav 32, 11–8 (1997). [DOI] [PubMed] [Google Scholar]
- Millar R. et al. A novel mammalian receptor for the evolutionarily conserved type II GnRH. Proc Natl Acad Sci U S A 98, 9636–41 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett D. K., Bunnell T. M., Millar R. P. & Abbott D. H. Gonadotropin-releasing hormone II stimulates female sexual behavior in marmoset monkeys. Endocrinology 147, 615–23 (2006). [DOI] [PubMed] [Google Scholar]
- Temple J. L., Millar R. P. & Rissman E. F. An evolutionarily conserved form of gonadotropin-releasing hormone coordinates energy and reproductive behavior. Endocrinology 144, 13–9 (2003). [DOI] [PubMed] [Google Scholar]
- Bhatnagar S., Vining C., Iyer V. & Kinni V. Changes in hypothalamic-pituitary-adrenal function, body temperature, body weight and food intake with repeated social stress exposure in rats. J Neuroendocrinol 18, 13–24 (2006). [DOI] [PubMed] [Google Scholar]
- Lenard N. R. & Berthoud H. R. Central and peripheral regulation of food intake and physical activity: pathways and genes. Obesity (Silver Spring) 16 Suppl 3, S11–22 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman A. S. & Rissman E. F. The evolutionarily conserved gonadotropin-releasing hormone II modifies food intake. Endocrinology 145, 686–91 (2004). [DOI] [PubMed] [Google Scholar]
- Abraham E., Palevitch O., Gothilf Y. & Zohar Y. The zebrafish as a model system for forebrain GnRH neuronal development. Gen Comp Endocrinol 164, 151–60 (2009). [DOI] [PubMed] [Google Scholar]
- Honjo Y., Payne L. & Eisen J. S. Somatosensory mechanisms in zebrafish lacking dorsal root ganglia. J Anat 218, 271–6 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W. & Delay R. J. Gonadotropin-releasing hormone modulates voltage-activated sodium current and odor responses in Necturus maculosus olfactory sensory neurons. J Neurosci Res 85, 1656–67 (2007). [DOI] [PubMed] [Google Scholar]
- Abraham E. et al. Early development of forebrain gonadotrophin-releasing hormone (GnRH) neurones and the role of GnRH as an autocrine migration factor. J Neuroendocrinol 20, 394–405 (2008). [DOI] [PubMed] [Google Scholar]
- Sharan S. K., Thomason L. C., Kuznetsov S. G. & Court D. L. Recombineering: a homologous recombination-based method of genetic engineering. Nat Protoc 4, 206–23 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warming S., Costantino N., Court D. L., Jenkins N. A. & Copeland N. G. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res 33, e36 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Page L. & Sherwood N. M. A role for GnRH in early brain regionalization and eye development in zebrafish. Mol Cell Endocrinol 257–258, 47–64 (2006). [DOI] [PubMed] [Google Scholar]
- Moeller J. F. & Meredith M. Differential co-localization with choline acetyltransferase in nervus terminalis suggests functional differences for GnRH isoforms in bonnethead sharks (Sphyrna tiburo). Brain Res 1366, 44–53 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zmora N. et al. The GnRH system in the European sea bass (Dicentrarchus labrax). J Endocrinol 172, 105–16 (2002). [DOI] [PubMed] [Google Scholar]
- Zmora N. et al. Differential and gonad stage-dependent roles of kisspeptin1 and kisspeptin2 in reproduction in the modern teleosts, morone species. Biol Reprod 86, 177 (2012). [DOI] [PubMed] [Google Scholar]
- Xia W., Zhou L., Yao B., Li C. J. & Gui J. F. Differential and spermatogenic cell-specific expression of DMRT1 during sex reversal in protogynous hermaphroditic groupers. Mol Cell Endocrinol 263, 156–72 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Supplementary Video 1
Supplementary Video 2