Abstract
Possible conformations for a cyclic nonapeptide that are consistent with conformation-dependent information obtained from an NMR investigation of the peptide in solution are presented. These several conformations are deduced from the myriad of possible conformations by eliminating from consideration all cyclic species having one or more residues in a conformation that does not correspond to the vicinal coupling constants observed by NMR between the amide and α-protons. A Karplus-like relation connecting the dihedral angle [unk]′ and the vicinal coupling JNα between N—H and Cα—Hα is used to test this correspondence. A further reduction in the number of cyclic conformations under consideration is made possible by rejecting the conformations that have a high intramolecular conformational energy. The intramolecular conformational energy of the cyclic nonapeptide is estimated by summing the independent residue energies. These have been calculated by others with approximate potential functions to account for the intrinsic torsional potentials and the nonbonded steric (6-12 potential) and electrostatic (monopole-monopole) interactions solely dependent upon one or both of the residue rotations, [unk] and Ψ, about the N—Cα and Cα—C bonds, respectively.
Full text
PDF
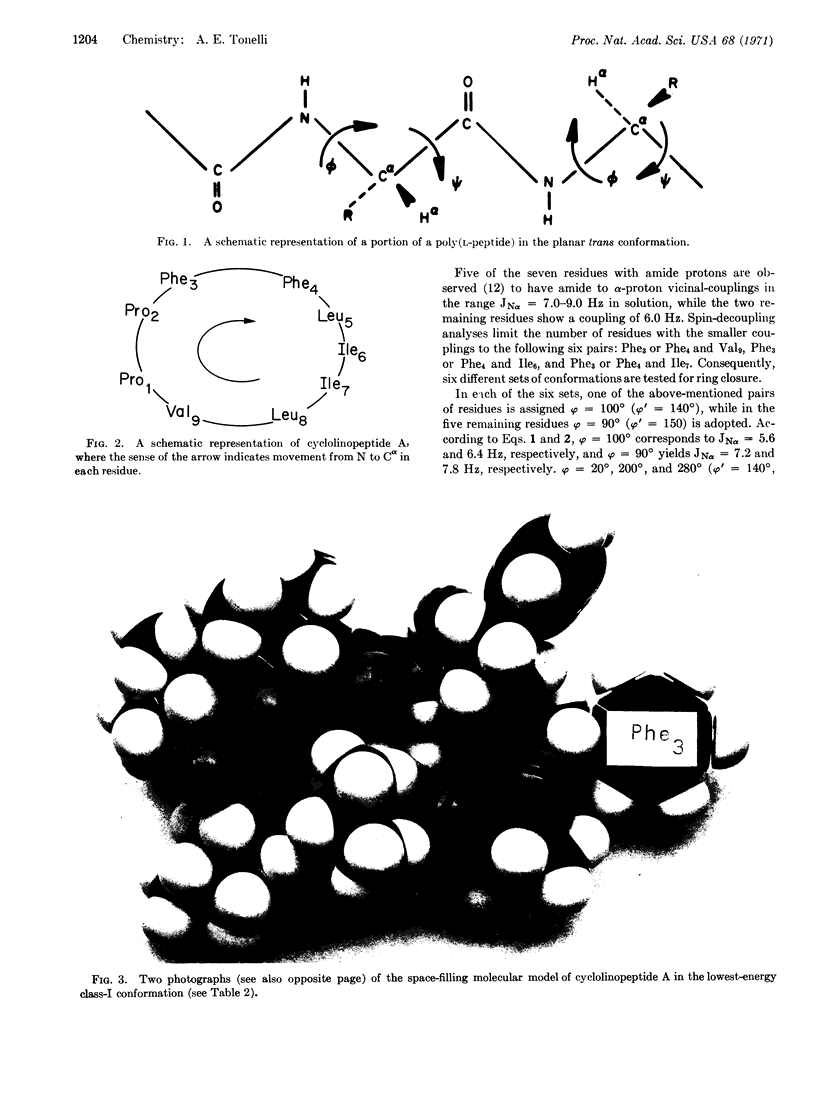
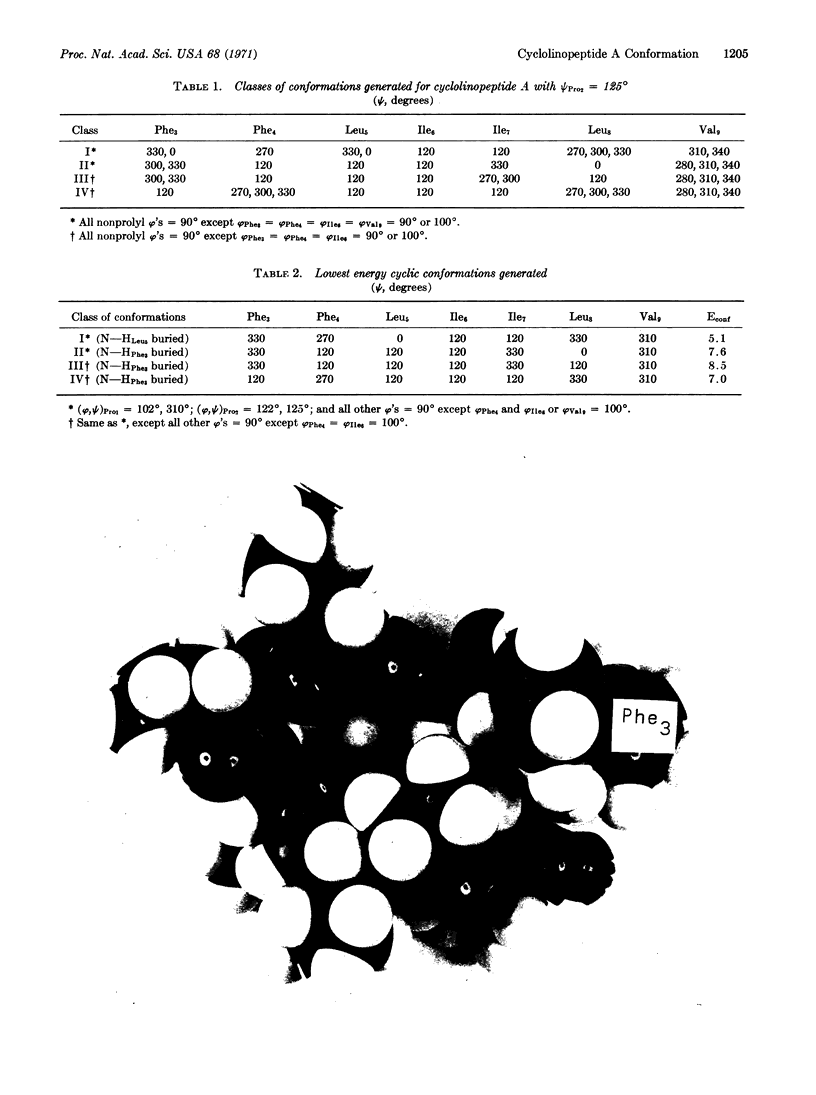


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brewster A. I., Bovey F. A. Conformation of cyclolinopeptide a observed by nuclear magnetic resonance spectroscopy. Proc Natl Acad Sci U S A. 1971 Jun;68(6):1199–1202. doi: 10.1073/pnas.68.6.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bystrov V. F., Portnova S. L., Tsetlin V. I., Ivanov V. T., Ovchinnikov Y. A. Conformational studies of peptide systems. The rotational states of the NH--CH fragment of alanine dipeptides by nuclear magnetic resonance. Tetrahedron. 1969 Feb;25(3):493–515. doi: 10.1016/s0040-4020(01)83261-0. [DOI] [PubMed] [Google Scholar]
- De Santis P., Giglio E., Liquori A. M., Ripamonti A. Van der Waals interaction and the stability of helical polypeptide chains. Nature. 1965 May 1;206(983):456–458. doi: 10.1038/206456a0. [DOI] [PubMed] [Google Scholar]
- IUPAC-IUB Commission on Biochemical Nomenclature. Abbreviations and symbols for the description of the conformation of polypeptide chains. Tentative rules (1969). Biochemistry. 1970 Sep 1;9(18):3471–3479. doi: 10.1021/bi00820a001. [DOI] [PubMed] [Google Scholar]
- Leach S. J., Némethy G., Scheraga H. A. Computation of the sterically allowed conformations of peptides. Biopolymers. 1966 Apr-May;4(4):369–407. doi: 10.1002/bip.1966.360040402. [DOI] [PubMed] [Google Scholar]
- Miller W. G., Goebel C. V. Dimensions of protein random coils. Biochemistry. 1968 Nov;7(11):3925–3935. doi: 10.1021/bi00851a021. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan C., Ramachandran G. N. Stereochemical criteria for polypeptide and protein chain conformations. II. Allowed conformations for a pair of peptide units. Biophys J. 1965 Nov;5(6):909–933. doi: 10.1016/S0006-3495(65)86759-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmel P. R., Flory P. J. Conformational energies and configurational statistics of copolypeptides containing L-proline. J Mol Biol. 1968 May 28;34(1):105–120. doi: 10.1016/0022-2836(68)90237-4. [DOI] [PubMed] [Google Scholar]
- Schimmel P. R., Flory P. J. Conformational energy and configurational statistics of poly-L-proline. Proc Natl Acad Sci U S A. 1967 Jul;58(1):52–59. doi: 10.1073/pnas.58.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]