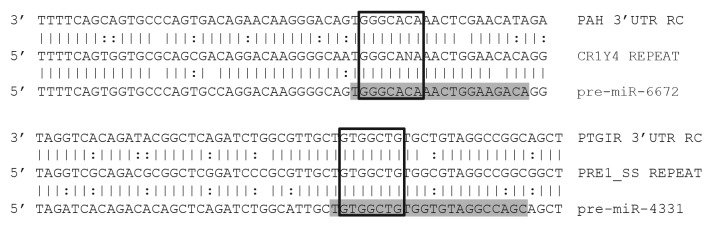
Figure 6. Alignments of miRs with predicted targets. Illustration shows a predicted miR target 3′UTRs on top, a consensus transposable element in the middle, and the corresponding miR sequence on the bottom. Mature miRs are highlighted in gray. Open boxes indicate perfect seed matches. To qualify as a 3′UTR match alignments were required to 1) contain a perfect seed match, 2) match ≥ 50% of the flanking sequence used in the target query, and 3) occur within a 3′UTR sequence aligning to a miR’s progenitor TE sequence. Vertical lines indicate base identity with the TE consensus sequence. Dotted lines indicate purine/pyrimidine conservation. PAH, Gallus gallus phenylalanine-4-hydroxylase ENSGALG00000012754. PTGIR, Sus scrufa prostaglandin I2 (prostacyclin) receptor ENSSSCG00000026602. RC, reverse complemented
