Abstract
The cytochromes P450 (CYPs) are heme proteins responsible for the oxidation of xenobiotics and pharmaceuticals and the biosynthesis of essential steroid products. In all cases, substrate binding initiates the enzymatic cycle, converting ferric low spin (LS) to high-spin (HS), with the efficiency of the conversion varying widely for different substrates, so documentation of this conversion for a given substrate is an important objective. Resonance Raman (rR) spectroscopy can effectively yield distinctive frequencies for the ν3 “spin state marker” bands. Here, employing a reference cytochrome P450 (CYP101), the intensities of the ν3 modes (ILS) and (IHS) relative to an internal standard (sodium sulfate) yield relative populations for the two spin states; i.e., a value of 1.24 was determined for the ratio of the relative cross sections for the ν3 modes. Use of this value was then shown to permit a reliable calculation of relative populations of the two spin states from rR spectra of several other Cytochromes P450. The importance of this work is that, using this information, it is now possible to conveniently document by rR the spin state population without conducting separate experiments requiring different analytical methods, instrumentation and additional sample.
Keywords: Cytochrome P450, Raman, spin state
The cytochromes P450 (CYPs) are heme-based monooxgenases responsible for the oxidative metabolisim of a huge number of relatively inert substrates, including pharmaceuticals and other xenobiotics, and for the biosynthesis of essential steroid products.[1-4] Substrate binding initiates the enzymatic cycle by triggering a crucial low spin (LS) to high spin (HS) state change that facilitates reduction to the ferrous heme intermediate that rapidly binds oxygen, which is then converted to the highly reactive Compound I. [5] Inasmuch as the efficiency of spin state conversion varies widely for different substrates and enzymes,[1,2,6,7] documentation of the extent of this conversion for a given substrate is an important experimental objective. Resonance Raman (rR) spectroscopy can effectively document the presence of HS or LS states of the ferric heme by appearance of distinctive frequencies observed at ~1485 and 1500 cm−1, respectively.[8-10] Given recent advances in producing and stabilizing these extremely important enzymes, [11-13] it is anticipated that applications of rR to these proteins will now expand considerably. The purpose of this work is to provide a systematic approach to utilize rR spectroscopy to reliably estimate spin state populations for different substrate/enzyme combinations.
Cytochrome P450cam (CYP101), an ideal reference protein, exhibits an almost complete spin state conversion upon binding its natural substrate, camphor, switching from 96% LS to 95% HS, as documented by electronic absorption spectrophotometry, Fig. S1 (Supporting Information).[14] The CYP101 was expressed and purified as published earlier;[11,15] experimental procedures, including sample preparation for rR measurements, are presented in Supporting Information. The laser excitation lines for these studies were 406.7 and 413.1 nm.[8-10, 16-19]
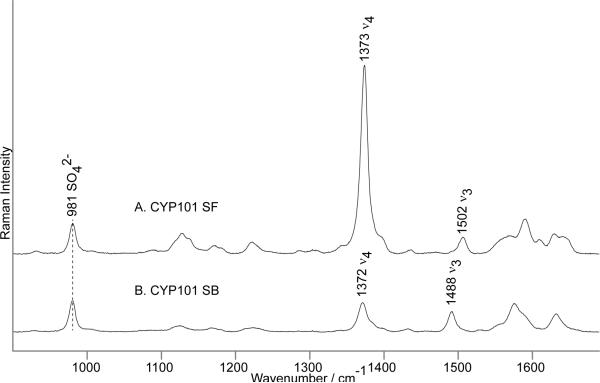
As shown in Figure 1, the spin state change is observed as a shift of ν3 from 1502 cm−1 (LS) to 1488 cm−1(HS). To estimate relative rR cross sections for the substrate-free (SF) and substrate-bound (SB) samples, three samples of these two forms, each containing 0.500 M internal standard Na2SO4, were measured and their spectra were analyzed using the following peak fitting procedure. The SO42- bands at 981 cm−1 (and all heme modes) were found to fit best with a 30% Gaussian/ 70% Lorentzian function. The average band width for the sulfate mode for all six spectra was 10.1 cm−1; the resulting band widths of the spin-marker bands were 11.5 cm−1 (1488 cm−1) and 11.3 cm−1 (1502 cm−1). Peak areas, instead of peak heights, were used to calculate the relative cross sections of the spin state marker bands; i.e., ILS/IIS and IHS/IIS, where IIS is the intensity of the 981 cm−1 band of sulfate. Noting that SF CYP101 is 96 % low spin and the camphor-bound is 95% high spin,[14] the operative relative intensities, YLS and YHS, values were derived by dividing the raw relative intensities by 0.96 and 0.95 factors, respectively. The YHS/YLS ratios of the ν3 bands were calculated for all nine combinations derived from six samples and the results are presented in Table S1 of Supporting Information. The YHS/YLS ratio for the ν3 mode with 406.7 nm excitation line is 1.24 ± 0.06; similar calculations for spectra measured with the 413.1 nm excitation line yielded a YHS/YLS ratio of 1.19 ± 0.04. Though of borderline significance, this smaller value is reasonable, because the 413.1 nm line is closer to resonance with the Soret band of the LS sample (417 nm) (Table S1, Supporting Information).
Figure 1.
The resonance Raman spectra of ferric CYP101 substrate-free (A) and substrate-bound (B). Spectra measured with 406.7 nm excitation line and normalized to the sulfate band at 981 cm−1.
In order to expand potential applications of this procedure, these ratios were also calculated for the ν4 and ν7 modes with both excitation lines (Table S2, Supporting Information). These data can be used to normalize spectra in different regions. One can apply the YHS/YLS ratio of 0.21 ± 0.013 for the intense ν4 mode when normalizing high frequency spectra (Fig. S2, Supporting Information) and in the low frequency region one could utilize the 0.38 ± 0.020 ratio for ν7 mode.
Given that the electronic spectra of both the HS and LS states of the bacterial CYPs correspond well with those of mammalian CYPs,[20] it is reasonable to expect that the value of 1.24 derived here for CYP101 should be valid for spectra of mammalian CYPs. To evaluate this issue, the derived 1.24 value was applied to calculate the percentage of spin state conversion upon substrate binding of several mammalian cytochromes available in our laboratory; i.e., CYP2B4, ND:CYP3A4 and ND:CYP17.[21-25] The percentages of LS and HS states calculated from rR spectra (406.7 nm excitation) using the method presented above were compared with percentages independently derived from available UV-Vis data (Table1). As can be seen, the data match quite well for CYP2B4 with butylated hydroxytoluene (BHT), CYP3A4 both SF and with erythromycin (ERY) as well as CYP17 with progesterone (PROG) and 17-hydroxyprogesterone (17-OH PROG). The value obtained here for the testosterone (TST)-bound ND:CYP3A4 falls within the range reported for measurements with UV-visible spectrophotometry; as is discussed more thoroughly in several earlier works dealing with substrate-binding equilibria and spin state conversion for these systems,[22-24] the percentage LS→ HS conversion is not linearly related to the number of bound substrates and is further complicated by the fact that substrate access to enzyme also depends on partitioning of the substrate into the lipid bilayer of the nanodisc. Similar complications can also be encountered for the bromocryptine (BC)-bound samples of the ND:CYP3A4 system and may account for the slightly larger, but not unreasonable, discrepancy seen here; i.e., 80% vs 93%.[26]
Table 1.
The calculated percentage of spin state populations in various cytochromes P450 measured with 406.7 nm excitation line and using the YHS/YLS ratio of 1.24 the v3 modes. The calculated data are compared with the data derived from UV-Vis spectra.
Supplementary Material
Acknowledgments
This work was supported in part by grants from the National Institutes of Health (GM96117) and the National Science Foundation (MCB 0951110) to JRK.
Footnotes
Supporting information
Supporting information may be found in the online version of this article.
References
- 1.Ortiz de Montellano PR, editor. Cytochrome P450: Structure, Mechanism, and Biochemistry. Kluwer Academic/Plenum Publisher; New York: 2005. [Google Scholar]
- 2.Sigel A, Sigel H, Sigel RKO, editors. Metal Ions in Life Sciences. Vol. 3. John Wiley & Sons, Ltd.; 2007. [Google Scholar]
- 3.Guengerich FP. Ann. Rev. Pharmacol. Toxicol. 1999;39:1. doi: 10.1146/annurev.pharmtox.39.1.1. [DOI] [PubMed] [Google Scholar]
- 4.Miller WL, Auchus RJ. Endocr. Rev. 2011;32:81. doi: 10.1210/er.2010-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Denisov IG, Makris TM, Sligar SG, Schlichting I. Chem. Rev. 2005;105:2253. doi: 10.1021/cr0307143. [DOI] [PubMed] [Google Scholar]
- 6.Sligar SG, Gunsalus IC. Proc. Natl. Acad. Sci. U.S.A. 1976;73:1078. doi: 10.1073/pnas.73.4.1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fisher MT, Sligar SG. J. Am. Chem. Soc. 1985;107:5018. [Google Scholar]
- 8.Spiro TG, editor. Biological Applicfations of Raman Spectroscopy. John Wiley & Sons; New York: 1988. [Google Scholar]
- 9.Kincaid JR. In: The Porphyrin Handbook. Kadish KM, Smith KM, Guilard R, editors. Vol. 7. Academic Press; 2000. p. 225. [Google Scholar]
- 10.Spiro TG, Soldatova AV, Balakrishnan G. Coord. Chem. Rev. 2013;257:511. doi: 10.1016/j.ccr.2012.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sibbesen O, Voss JJ, Ortiz de Montellano PR. J. Biol. Chem. 1996;271:22462. doi: 10.1074/jbc.271.37.22462. [DOI] [PubMed] [Google Scholar]
- 12.Denisov IG, Sligar SG. Biochim. Biophys. Acta. 2011;1814:223. doi: 10.1016/j.bbapap.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Denisov IG, Mak PJ, Makris TM, Sligar SG, Kincaid JR. J. Phys. Chem. A. 2008;112:13172. doi: 10.1021/jp8017875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fisher MT, Sligar SG. Biochemistry. 1985;24:6696. doi: 10.1021/bi00344a059. [DOI] [PubMed] [Google Scholar]
- 15.O'Keeffe DH, Ebel RE, Peterson JA. Methods in Enzymology. 1978;52:151. doi: 10.1016/s0076-6879(78)52017-x. [DOI] [PubMed] [Google Scholar]
- 16.Mak PJ, Kaluka D, Manyumwa EM, Zhang H, Deng T, Kincaid JR. Biopolymers. 2008;89:1045. doi: 10.1002/bip.21058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wells AV, Li P, Champion PM, Martinis SA, Sligar SG. Biochemistry. 1992;31:4384. doi: 10.1021/bi00133a002. [DOI] [PubMed] [Google Scholar]
- 18.Tosha T, Kagawa N, Arase M, Waterman MR, Kitagawa T. J. Biol. Chem. 2008;283:3708. doi: 10.1074/jbc.M707338200. [DOI] [PubMed] [Google Scholar]
- 19.Mak PJ, Denisov IG, Grinkova YV, Sligar SG, Kincaid JR. J. Am. Chem. Soc. 2011;133:1357. doi: 10.1021/ja105869p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sato R, Omura T. Cytochrome P-450. Academic Press; New York: 1978. [Google Scholar]
- 21.Personal communication. Dr. Sang-Choul Im; Department of Anesthesiology, University of Michigan.
- 22.Das A, Grinkova YV, Sligar SG. J. Am. Chem. Soc. 2007;129:13778. doi: 10.1021/ja074864x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frank DJ, Denisov IG, Sligar SG. J. Biol. Chem. 2011;286:5540. doi: 10.1074/jbc.M110.182055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Denisov IG, Baas BJ, Grinkova YV, Sligar SG. J. Biol. Chem. 2007;282:7066. doi: 10.1074/jbc.M609589200. [DOI] [PubMed] [Google Scholar]
- 25.Personal communication. Michael Gregory, University of Illinois at Urbana Champaign.
- 26.Nath A, Grinkova YV, Sligar SG, Atkins WM. J. Biol. Chem. 2007;282:28309. doi: 10.1074/jbc.M703568200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.