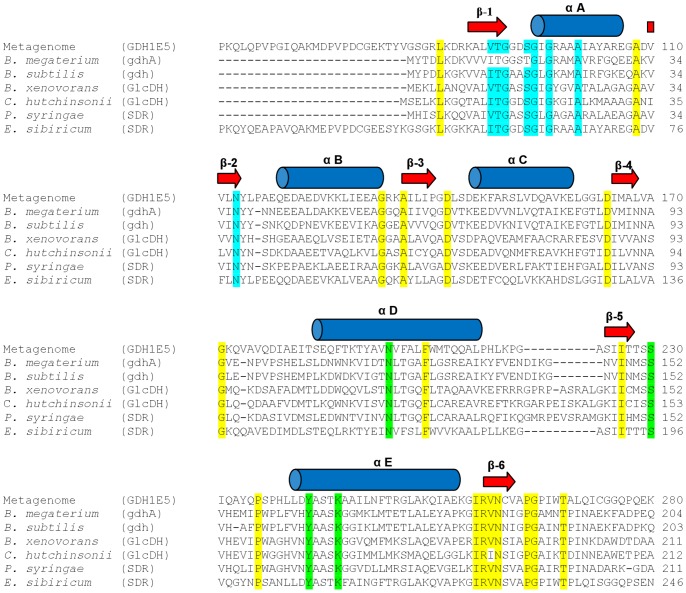
Figure 1. Amino acid sequence alignment of the GDH1E5 with several glucose dehydrogenases and SDR enzymes.
The secondary structure of the GDH1E5 (CCK35875) was predicted by the online tool PSIPRED 3.0 and is indicated as arrows for β-strands and as cylinders for α-helices. Conserved structural residues of SDR enzymes are highlighted. Active site residues are colored in green, residues for co-factor binding in blue and conserved structural residues in yellow. The sequence of the GDH1E5 was aligned with Glucose 1-Dehydrogenases (gdhA, gdh, GlcDH) from Bacillus megaterium (P10528), Bacillus subtilis (P12310), Burkholderia xenovorans (YP_554854), Cytophaga hutchinsonii (YP_678271), and two additional short-chain dehydrogenases (SDR) from Pseudomonas syringae (YP_235378) and Exiguobacterium sibiricum (YP_001815303).