Summary
Avian influenza A viruses rarely infect humans, but if they do and transmit among them, worldwide outbreaks (pandemics) can result. The recent sporadic infections of humans in China with a previously unrecognized avian influenza A virus of the H7N9 subtype (A(H7N9)) have caused concern due to the appreciable case fatality rate associated with these infections (>25%), potential instances of human-to-human transmission1, and the lack of pre-existing immunity among humans to viruses of this subtype. Here, we therefore characterized two early human A(H7N9) isolates, A/Anhui/1/2013 and A/Shanghai/1/2013 (H7N9; hereafter referred to as Anhui/1 and Shanghai/1, respectively). In mice, Anhui/1 and Shanghai/1 were more pathogenic than a control avian H7N9 virus (A/duck/Gunma/466/2011; H7N9; Dk/GM466) and a representative pandemic 2009 H1N1 virus (A/California/04/2009; H1N1; CA04). Anhui/1, Shanghai/1, and Dk/GM466 replicated well in the nasal turbinates of ferrets. In nonhuman primates (NHPs), Anhui/1 and Dk/GM466 replicated efficiently in the upper and lower respiratory tracts, whereas the replicative ability of conventional human influenza viruses is typically restricted to the upper respiratory tract of infected primates. By contrast, Anhui/1 did not replicate well in miniature pigs upon intranasal inoculation. Most critically, Anhui/1 transmitted via respiratory droplets in one of three pairs of ferrets. Glycan arrays demonstrated that Anhui/1, Shanghai/1, and A/Hangzhou/1/2013 (a third human A(H7N9) virus tested in this assay) bind to human virus-type receptors, a property that may be critical for virus transmissibility in ferrets. Anhui/1 was less sensitive than a pandemic 2009 H1N1 virus to neuraminidase inhibitors, although both viruses were equally susceptible to an experimental antiviral polymerase inhibitor. The robust replicative ability in mice, ferrets, and NHPs and the limited transmissibility in ferrets of Anhui/1 suggest that A(H7N9) viruses have pandemic potential.
Influenza A virus infections place a considerable burden on public health and the world economy. In March, 2013, several individuals were reported to be infected with an avian A(H7N9) virus1,2. Viruses of this subtype do not circulate in humans, so A(H7N9) viruses capable of transmitting among humans would encounter populations that lack any protective immunity to them. By May 30, 2013, 132 confirmed human infections with A(H7N9) viruses had been reported with 37 deaths (http://www.who.int/influenza/human_animal_interface/influenza_h7n9/08_ReportWebH7N9Number.pdf), resulting in a case fatality rate of >25%.
Sequence and phylogenetic analysis revealed that the haemagglutinin (HA) and neuraminidase (NA) genes of the A(H7N9) viruses originated from avian H7 and N9 viruses, respectively2–4, whereas the remaining six genes are closely related to H9N2 subtype viruses that have circulated in poultry in China2–4. Several of the A(H7N9) viruses possess amino acid changes known to facilitate infection of mammals, such as leucine at position 226 of HA (H3 HA numbering), which confers increased binding to human-type receptors5, and the mammalian-adapting mutations E627K6,7 or D701N8 in the PB2 polymerase subunit. Notably, the PB2-627K or PB2-701N markers have been detected in almost all human, but not avian or environmental A(H7N9) isolates, suggesting ready adaptation of A(H7N9) viruses to humans.
To characterize the biological properties and pandemic potential of A(H7N9) viruses, we compared Anhui/1 (which possesses the mammalian-adapting HA-226L and PB2-627K markers) and Shanghai/1 (which possesses the ‘avian-type’ HA-226Q and mammalian-adapting PB2-627K markers) with the phylogenetically unrelated avian H7N9 Dk/GM466 virus, and with CA04, an early, representative 2009 H1N1 pandemic virus. Anhui/1, Shanghai/1, and CA04 replicated efficiently in Madin–Darby canine kidney (MDCK) and in differentiated human bronchial epithelial (NHBE) cells compared with Dk/GM466, especially at 33°C, a temperature corresponding to the human upper airway (Figure S1). Electron microscopic analysis showed Anhui/1 as a spherical particle that appeared to be efficiently released from infected cells (Figure S2).
Next, we assessed the pathogenicity of Anhui/1 and Shanghai/1 in established animal models in influenza virus research, namely mice, ferrets, and NHPs (Anhui/1 only). In BALB/c mice, Anhui/1 and Shanghai/1 were more pathogenic than CA04 and Dk/GM466 based on MLD50 (mouse lethal dose 50; the dose required to kill 50% of infected mice) values, which were 103.5 plaque-forming units (PFU) for Anhui/1 and Shanghai/1, 105.5 PFU for CA04, and 106.7 PFU for Dk/GM466 (Figure S3). Three days post-infection (dpi), virus titres in the lungs and nasal turbinates of Anhui/1-, Shanghai/1- and CA04-infected mice were slightly higher than those in Dk/GM466-infected mice (Table S1). Lung lesions in Anhui/1- and CA04-infected mice were more severe than those in Dk/GM466-infected mice, in particular on 6 dpi (Figure S4). Bronchitis, bronchiolitis, thickening of the alveolar septa, edema, and interstitial inflammatory cell infiltration were also more prominent in Anhui/1- and CA04-infected mice. Viral antigen was detected in many alveolar and bronchial epithelial cells at 3 dpi in Anhui/1- and CA04-infected mice (Figure S4), whereas viral antigen-positive cells were restricted to a few bronchial epithelial cells in Dk/GM466-infected mice (Figure S4). Collectively, these findings demonstrate that Anhui/1 is as pathogenic as CA04 and more pathogenic than Dk/GM466 in mice.
Ferrets intranasally infected with Anhui/1, Shanghai/1, CA04, or Dk/GM466 experienced loss of appetite. Transient weight loss was detected in one of the three animals infected with Anhui/1 (Figure S5). Virus titres in the trachea of Anhui/1-, Shanghai/1- and CA04-infected ferrets were higher at 3 dpi than those obtained from Dk/GM466-infected animals (Table S2); by 6 dpi, virus was isolated from the trachea of Anhui/1-, Shanghai/1- and Dk/GM466-infected animals, but not from that of CA04-infected ferrets. All three viruses replicated inefficiently in the lungs of these animals. In this study, we did not detect virus in the lungs in the CA04- infected animals at 3 dpi (Table S2), whereas this virus was recovered from two of three ferrets infected at 3dpi in our previous study9, consistent with efficient replication of pandemic 2009 H1N1 virus in the lungs of ferrets as reported by others10–13. Appreciable amounts of virus were recovered from nasal turbinates at 3 and 6 dpi, with the exception of the nasal turbinates of CA04-infected animals at 6 dpi (Table S2). Inflammation was prominent in the trachea and submucosal glands of CA04-infected ferrets (Figure S6). Viral antigen-positive cells were detected in the tracheal, glandular, and alveolar epithelial cells of all ferrets. Anhui/1- and CA04- infected ferrets displayed numerous viral antigen-positive cells especially in the glandular epithelia, whereas Dk/GM466-infected ferrets presented far fewer antigen-positive cells. Viral antigens were also detected in pneumocytes in localized lung lesions of each ferret. In mediastinal lymph nodes, viral antigen was detected in only Anhui/1- and CA04-infected ferrets. Anhui/1 and Shanghai/1 thus established a robust, though relatively mild infection in the upper respiratory organs of ferrets that was unlike most avian H5N1 influenza virus infections in ferrets, which consistently cause severe symptoms including profound weight loss.
Infection of cynomolgus macaques (Macaca fascicularis) with 6.7 ×107 PFU of Anhui/1 or Dk/GM466 caused fever (Figure S7), as did infection with CA04 in our previous study9. Both Anhui/1 and Dk/GM466 replicated appreciably in macaque nasal turbinates, trachea, and lungs, although variability among the virus titres was noticed, as is commonly found among outbred animals (Table S3, S4a). Previously, we detected efficient replication of CA04 in the respiratory organs of cynomolgus macaque at 3 dpi; by 7 dpi, no virus was detected in the lungs9.
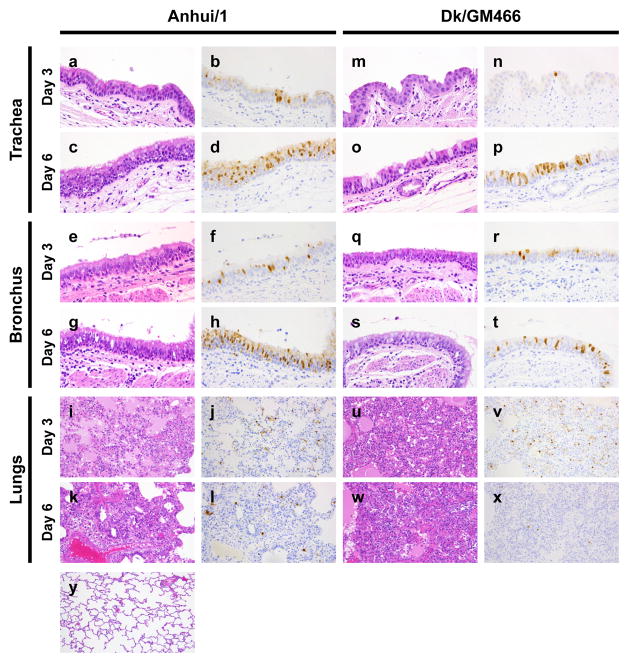
Pathological examination of the respiratory organs did not reveal major differences between macaques infected with Anhui/1 or Dk/GM466; no remarkable lesions were detected in the trachea or lobular bronchus, but alveolar spaces contained edematous exudate and inflammatory infiltrates comprising mainly neutrophils and monocytes/macrophages (Figure 1). At 6 dpi, regenerative changes were observed. Lung inflammation scores did not reveal appreciable differences between animals infected with Anhui/1 or Dk/GM466 (Table S4b). Numerous tracheal and bronchial epithelial cells of Anhui/1-infected macaques were positive for viral antigen (representative images of the areas that stained most intensely with the anti-NP antibody are shown for animals #3, #6, #8, and #10 in Figure 1), as was observed in our previous study with CA04 at 7 dpi9. Fewer antigen-positive cells in the tracheal and bronchial epithelia of Dk/GM466-infected animals were detected in particular at 3 dpi (Figure 1). Viral antigen was also detected in the mediastinal lymph node sections of Anhui/1-infected monkeys (Figure S8). In addition, our analysis of chemokine/cytokine responses suggests that Anhui/1 induces strong inflammatory responses both systemically and at the site of virus infection (Figure S9 and Supplementary Information).
Figure 1. Pathological findings in infected macaques.
Shown are pathological findings in the trachea (a–d, m–p), bronchus (e–h, q–t), and lungs (i–l, u–x) of macaques infected with Anhui/1 (a–l) or Dk/GM466 (m–x) at 3 dpi (a, b, e, f, i, j, m, n, q, r, u, v) or 6 dpi (c, d, g, h, k, l, o, p, s, t, w, x) with HE staining (a, c, e, g, i, k, m, o, q, s, u, w) or immunohistochemistry for influenza viral antigen (b, d, f, h, j, l, n, p, r, t, v, x). HE staining of the lung of an uninfected macaque is shown (y). Original magnification: x 400 (a–h, m–t), x 200 (i–l, u–y).
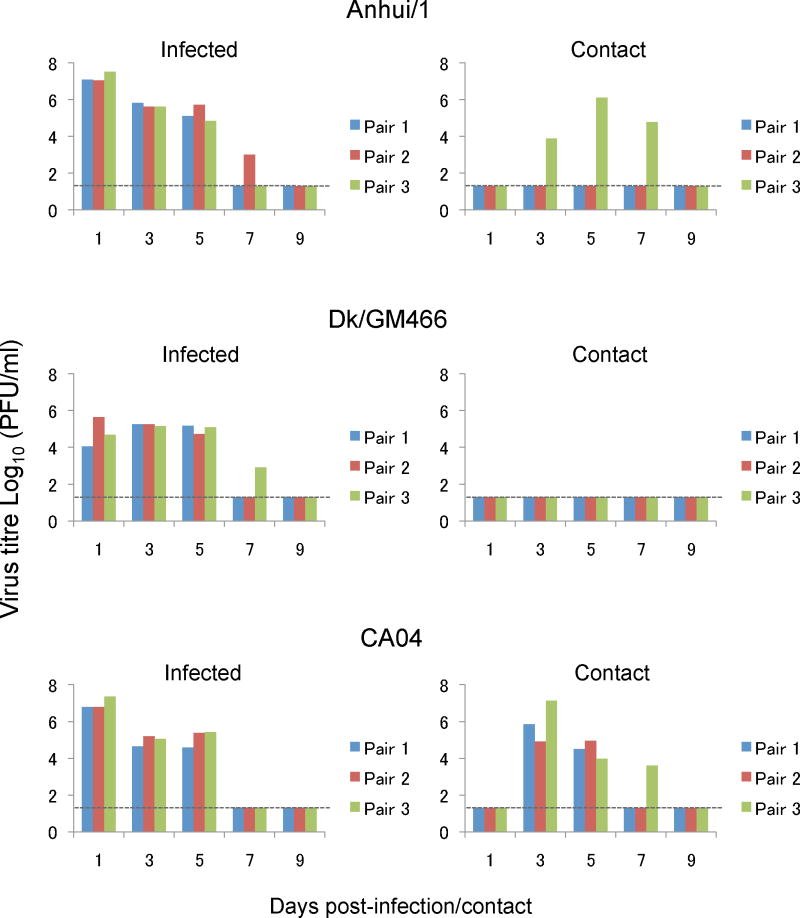
No sustained human-to-human transmission of the novel A(H7N9) viruses has been reported to date. To assess the transmissibility of Anhui/1, naïve ‘contact’ ferrets in wireframe cages (which prevent direct contact with animals in neighbouring cages, but allow respiratory droplet transmission) were placed adjacent to an infected ferret the day after infection as described previously14. We recovered viruses from the nasal washes of all three contact ferrets for CA04 (Figure 2), as expected based on previous studies by us9 and others12,13. No virus was detected in the nasal washes of contact ferrets for Dk/GM466 (Figure 2), consistent with the general lack of transmissibility of avian influenza viruses in ferrets. However, one of three contact ferrets for Anhui/1 shed virus on days 3–7 after contact (Figure 2). Serum antibody titres against Anhui/1 confirmed infection of this animal, whereas the other two contact animals did not seroconvert (Table S5). Given that avian H5N1 influenza viruses require several mutations to transmit via respiratory droplets among ferrets14–17, the pandemic potential of A(H7N9) viruses may be greater than that of the highly pathogenic avian H5N1 influenza viruses.
Figure 2. Respiratory droplet transmission in ferrets.
Ferrets were infected with 5 ×105 PFU of Anhui/1, Dk/GM466, or CA04 (inoculated ferrets). One day later, three naïve ferrets (contact ferrets) were each placed in a cage adjacent to an infected ferret. Nasal washes were collected from infected ferrets on day 1 after inoculation and from contact ferrets on day 1 after co-housing, and then every other day (up to 9 days) for virus titration.
Since replication and transmission of an H5 HA-possessing virus in ferrets is associated with amino acid changes in HA14,17, we sequenced the genomes of viruses obtained from infected and contact ferrets. Compared to virus inoculum, we detected three non-synonymous mutations in HA (T71I, R131K, and A135T; Figure 3; Table S6) and one non-synonymous mutation in NA (A27T; N9 numbering) (Table S6); we also detected several synonymous nucleotide changes. The amino acid mutations in HA were detected in >40% of the molecular clones derived from samples obtained on day 3 post-contact (Table S7), but were found exclusively by day 5 post-contact. Interestingly, the egg-grown virus stock of Anhui/1 possessed a mixture of amino acids at five positions, and the three amino acid changes in HA detected in the transmitted virus were detected in 2 of 39 HA clones representing the egg-grown virus stock of Anhui/1 (Table S7). The selection of these mutations during A/Anhui replication and/or transmission in ferrets strongly suggests that these mutations have a biological role, possibly in HA stability and/or receptor-binding specificity or affinity. In fact, positions 131 and 135 are located near the receptor-binding pocket (Figure 3). At position 71, both threonine and isoleucine are commonly found among H7 HAs, and the location of this position ‘underneath’ the receptor-binding pocket suggests a possible effect on HA stability (Figure 3).
Figure 3. Glycan microarray analysis and HA structural analysis.
(a) Localization of amino acid changes in a virus from a ferret infected via respiratory droplets. Shown is the three-dimensional structure of A/Netherlands/219/2003 (H7N7) HA (PDB ID: 4DJ6) in complex with human receptor analogues. (b) Close-up view of the globular head. Mutations that increase affinity to human-type receptors are shown in cyan. Mutations that emerged in Anhui/1 HA during replication and/or transmission in ferrets are shown in red (T71I), green (A135T), and blue (R131K). The human receptor analogue [derived from its complex with H9 HA (PDB ID: 1JSI); shown in orange] is docked into the structure. Images were created with MacPymol [http://www.pymol.org/]. (c–g) Receptor specificities of recombinant viruses possessing A(H7N9) HAs (Anhui/1, Shanghai/1, Hangzhou/1) were compared with representative avian (Vietnam/1203) and human (Kawasaki/173) isolates in a glycan microarray containing α2–3 and α2–6 sialosides. Error bars represent standard deviations calculated from 6 replicate spots of each glycan. A complete list of glycans is found in Table S9.
Our data indicate that Anhui/1 efficiently infects mammalian cells. We speculated that amino acid changes in Anhui/1 HA contribute to this host tropism. The HA-226L residue found in Anhui/1 is known to increase the affinity of H3 HAs (which are phylogenetically closely related to H7 HAs) to sialic acids linked to galactose by an α2–6-linkage (‘human-type’ receptors)5; by contrast, avian influenza viruses preferentially bind to sialic acids linked to galactose by an α2–3-linkage (‘avian-type’ receptors). In addition, Anhui/1 HA deviates from the avian virus consensus sequence at positions 186 and 189, which influence the receptor-binding preferences of H5 and H9 HAs, respectively18–21.
To analyze receptor-binding preference, we subjected recombinant viruses possessing the Anhui/1, Shanghai/1, or Hangzhou/1 HA genes (see Table S8) in combination with Anhui/1 NA genes and the remaining genes from A/Puerto Rico/8/34 (PR8; H1N1; a laboratory-adapted strain) to glycan array analysis. All three viruses bound to α2–6 linked sialosides, unlike a representative avian virus (A/Vietnam/1203/2004; H5N1, Vietnam/1203), which showed typical specificity for α2–3 linked sialosides (Figure 3; Table S9). Anhui/1 and Hangzhou/1 HAs bound most strongly to α2–6 linked sialosides, and in particular to extended N-linked glycans that are found on human bronchial epithelial cells22 (Figure 3). This binding pattern may be influenced by the ‘human-type’ residues at position 226 (Anhui/1 possesses HA-226L and Hangzhou/1 possesses HA-226I, the residues also found in human H3N2 influenza viruses at this position). Shanghai/1 HA (encoding the ‘avian-type’ HA-226Q) was less selective, binding equally well to both α2–6 and α2–3 linked sialosides (Figure 3). The specificity of the recombinant Anhui/1 and Hangzhou/1 HA viruses was comparable to the ‘human-type’ receptor specificity of H5N1 virus receptor mutants that exhibit respiratory droplet transmission in ferrets14,17, and to that of the human H7N3 A/New York/107/2003 isolate that exhibits contact, but not respiratory droplet transmission in ferrets23. Notably, however, inhibition of the NA enzymatic function by inclusion of the neuraminidase inhibitor zanamivir in the glycan array analysis resulted in substantial increases in binding to α2–3 linked sialosides (as shown for Hangzhou/1 in Figure S10). Moreover, preliminary analysis of the specificity of the recombinant Anhui/1 H7 HA revealed preferential binding to α2–3 sialosides (de Vries, R. P., McBride, R. and Paulson, J. C., unpublished observations). Thus, the ‘human-type’ receptor specificity of A(H7N9) viruses assessed by glycan array appears to reflect the combined activities of HA and NA.
In pigs, we observed a mild infection with no clinical symptoms (Table S10, S11, Figure S11, and Supplementary Information). Signs of disease were also absent in infected chickens and quails, which supported virus replication in a limited number of organs (a characteristic of low pathogenic avian influenza viruses) (Tables S12–15, and Supplementary Information).
Antiviral compounds are currently the only therapeutic and prophylactic option for A(H7N9) infections. We, therefore, determined the in vitro 50% inhibitory concentration (IC50) of several NA inhibitors (oseltamivir, zanamivir, laninamivir, and peramivir), and of an experimental inhibitor of the viral RNA polymerase (favipiravir, also known as T-705) against egg-grown virus stocks of Anhui/1 and Shanghai/1. Sanger sequencing of the Shanghai/1 NA gene revealed the R292K mutation known to confer resistance to NA inhibitors for N2 and N9 NAs24. Both Anhui/1 and Shanghai/1 were sensitive to all NA inhibitors tested (Table S16), consistent with a recent report that Shanghai/1 is susceptible to oseltamivir and zanamivir25, but inconsistent with the presence of the R292K mutation. A possible explanation for this discrepancy is that the Shanghai/1 isolate contained a mixture of drug-sensitive and -resistant NA genes in which the drug-sensitive subpopulation may not have been detected, as described by Wetherall et al.26. In fact, plaque purification of the egg-grown Shanghai/1 virus stock revealed a mixed population of NA genes encoding NA-292R or -292K. Testing of these variants confirmed that Shanghai/1-NA292R was susceptible to NA inhibitors, whereas the NA-292K variant was not (Table S16). Further testing demonstrated that 30% of oseltamivir-sensitive virus in the mixture is sufficient for results to be consistent with a purely sensitive virus population (Table S17). The IC50 values of favipiravir, determined by plaque reduction assays in MDCK cells, were low for Anhui/1 and the control CA04 virus (1.4 μg/ml and 1.2 μg/ml, respectively), suggesting that this compound could be a treatment option against A(H7N9) viruses resistant to NA inhibitors.
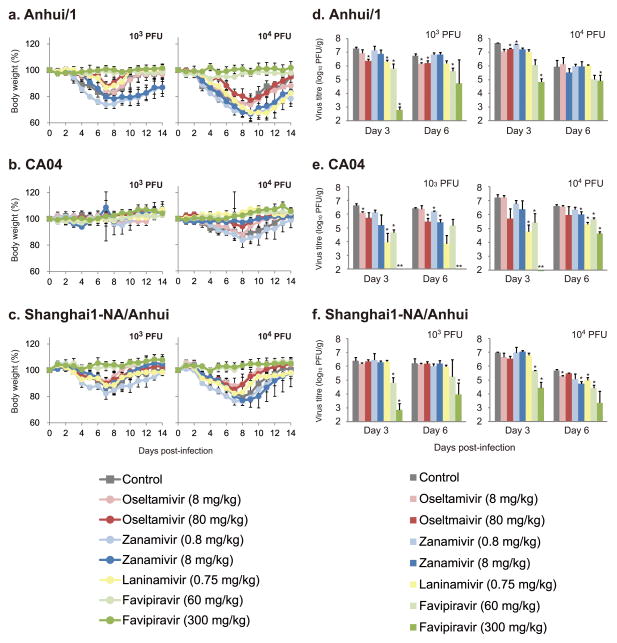
We also evaluated the therapeutic efficacy of the anti-influenza drugs in mice infected with Anhui/1, CA04, or a recombinant virus possessing the Shanghai/1 NA gene (encoding NA- 292K) with the remaining genes from Anhui/1. Peramivir, which is structurally similar to oseltamivir but is administered intravenously, was omitted from these experiments. Mice infected with 103 and 104 PFU of viruses were treated with the drugs beginning at 2 h post- infection. Some NA inhibitors had modest effects on the body weight loss of Anhui/1-infected mice (Figure 4a–c), consistent with limited, although statistically significant, effects on virus titre reduction (Figure 4d–f). We currently do not know if neuraminidase-resistant variants arose during Anhui/1 replication in mice, which may have limited virus susceptibility to NA inhibitors. In this context, it is interesting to note the poor efficacy of oseltamivir when used to treat a person infected with A(H7N9)27, presumably due to the emergence of drug-resistant variants. By contrast, favipiravir, which targets the viral polymerase complex, showed clear therapeutic effectiveness against both viruses at both doses tested.
Figure 4. Virus sensitivity to antivirals in mice.
Mice were intranasally inoculated with 103 or 104 PFU (50 μl) of Anhui/1 (a, d), CA04 (b, e), or recombinant Anhui/1 virus possessing Shanghai/1-NA-292K (c, f). At 2 h after infection, mice were treated with oseltamivir phosphate, zanamivir, laninamivir octanoate, favipiravir, PBS, or distilled water. Body weights were monitored daily (a–c). Three mice per group were euthanized at 3 and 6 dpi, and the virus titres in lungs were determined by plaque assays in MDCK cells (d–f). Statistically significant differences between virus titres of control mice and those of mice treated with antiviral drugs were determined by using Welch’s t-test or Student’s t-test on the result of the F-test. The resulting p-values were corrected by using Holm’s method (asterisk, p<0.05). Mean titres, or individual titres when virus was not recovered from all three animals (double-asterisk), are shown.
In summary, Anhui/1 and Shanghai/1 originated from an avian host, based on their sequences and phylogenetic relationships, yet possess several characteristic features of human influenza viruses, such as efficient binding to human-type receptors, efficient replication in mammalian cells (likely conferred by PB2-627K), and respiratory droplet transmission in ferrets (Anhui/1). These properties, together with the low efficacy of NA inhibitors and the lack of human immunity (we tested 500 human sera collected from various age groups in Japan and found no antibodies that recognized Anhui/1) make A(H7N9) viruses a formidable threat to public health.
Methods
Viruses
A/Anhui/1/2013 (H7N9; Anhui/1) and A/Shanghai/1/2013 (H7N9; Shanghai/1), both kindly provided by Dr. Yuelong Shu (Director of the WHO Collaborating Center for Reference and Research on Influenza, Director of the Chinese National Influenza Center, Deputy Director of the National Institute for Viral Disease Control and Prevention China CDC, Beijing, P. R. China), and A/duck/Gunma/466/2011 (H7N9; Dk/GM466) were propagated in embryonated chicken eggs. For antigenic characterization by haemagglutination inhibition (HI) assays (see below for detailed procedures), Anhui/1 was also propagated in Madin-Darby canine kidney (MDCK) cells. A/California/04/2009 (H1N1; CA04), A/Kawasaki/173/2001 (H1N1), and A/Vietnam/1203/2004 (H5N1) were propagated in MDCK cells. All experiments with H7N9 viruses were performed in enhanced biosafety level 3 (BSL3) containment laboratories at the University of Tokyo (Tokyo, Japan) and National Institute of Infectious Diseases, Japan, which are approved for such use by the Ministry of Agriculture, Forestry and Fisheries, Japan; or in enhanced BSL3 containment laboratories at the University of Wisconsin-Madison, which are approved for such use by the Centers for Disease Control and Prevention and by the US Department of Agriculture, USA.
Cells
MDCK cells were maintained in Eagle’s minimal essential medium (MEM) containing 5% newborn calf serum (NCS). Human embryonic kidney 293T cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal calf serum (FCS). Normal human bronchial epithelial cells (NHBE) were obtained from Lonza (Walkersville, MD). The monolayers of NHBE cells were cultured and differentiated as previously described28. All cells were incubated at 37°C with 5% CO2.
Antiviral compounds
Laninamivir and laninamivir octanoate were kindly provided by Daiichi Sankyo Co. Ltd. (Tokyo, Japan). Favipiravir was kindly provided by Toyama Chemical Co. Ltd. (Toyama, Japan) and oseltamivir carboxylate was provided by F. Hoffmann-La Roche Ltd. (Basel, Switzerland). Zanamivir was kindly provided by GlaxoSmithKline (Stevenage, UK). Peramivir was kindly provided by Shionogi & Co. Ltd. (Osaka, Japan).
Reverse genetics
Plasmid-based reverse genetics for influenza virus generation was performed as previously described29. Briefly, plasmids encoding the cDNAs for the eight viral RNA segments under the control of the human RNA polymerase I promoter and the mouse RNA polymerase I terminator (referred to as PolI plasmids), and plasmids for the expression of the viral PB2, PB1, PA, and NP proteins derived from a laboratory-adapted influenza A virus strain A/WSN/33 (H1N1) under the control of the chicken β-actin promoter30 were transfected into 293T cells with the help of a transfection reagent, Trans-IT 293 (Mirus, Madison, WI). At 48 h post-transfection, culture supernatants were harvested and inoculated to embryonated chicken eggs for virus propagation.
Growth kinetics of virus in cell culture
MDCK cells were infected in triplicate in 12-well plates with Anhui/1, Dk/GM466, or CA04 at a multiplicity of infection (MOI) of 0.01. After incubation at 37°C for 1 h, the viral inoculum was replaced with MEM containing 0.3% bovine serum albumin (with 0.75 μg/ml TPCK-treated trypsin), followed by further incubation at 37°C. Culture supernatants collected at the indicated time points were subjected to virus titration by using plaque assays in MDCK cells.
Cultures of differentiated NHBE cells were washed extensively with DMEM to remove accumulated mucus and infected in triplicate in 12-well plates with virus at an MOI of 0.001 from the apical surface. The inoculum was removed after 30 min of incubation at 33°C or 37°C, and cells were further incubated at 33°C or 37°C. Samples were collected at 6, 12, 24, 48, 72, and 96 h post-infection from the apical surface. Apical harvesting was performed by adding 500 μl of medium to the apical surface, followed by incubation for 30 min at 33°C or 37°C, and removal of the medium from the apical surface. The titres of viruses released into the cell culture supernatant were determined by plaque assay in MDCK cells.
Animal experiments
The sample sizes (n=3) for the mouse, ferret, quail, and chicken studies were chosen because they have previously been shown to be sufficient to evaluate a significant difference among groups 9,13,31–33. For the NHP and pig experiments, two or three animals per group were used and no statistical analysis was performed. No method of randomization was used to determine how animals were allocated to the experimental groups and processed in this study. The investigator was not blinded to the group allocation during the experiments or when assessing the outcome.
Experimental infection of mice
Six-week-old female BALB/c mice (Japan SLC Inc., Shizuoka, Japan) were used in this study. Baseline body weights were measured before infection. Under anaesthesia, four mice per group were intranasally inoculated with 101, 102, 103, 104, 105, or 106 plaque-forming units (PFU) (50 μl) of Anhui/1, Shanghai/1, Dk/GM466, or CA04. Body weight and survival were monitored daily for 14 days. For virological and pathological examinations, six mice per group were intranasally infected with 104 or 106 PFU (50 μl) of the viruses and three mice per group were euthanized at 3 and 6 days post-infection (dpi). The virus titres in various organs were determined by plaque assays in MDCK cells. All experiments with mice were performed in accordance with the University of Tokyo’s Regulations for Animal Care and Use and approved by the Animal Experiment Committee of the Institute of Medical Science, the University of Tokyo.
Experimental infection of ferrets
Five- to eight-month-old female ferrets (Triple F Farms, Sayre, PA), which were serologically negative by HI assay for currently circulating human influenza viruses, were used in this study. Under anaesthesia, six ferrets per group were intranasally inoculated with 106 PFU (0.5 ml) of Anhui/1, Shanghai/1, Dk/GM466, or CA04. Three ferrets per group were euthanized at 3 and 6 dpi for virological and pathological examinations. The virus titres in various organs were determined by plaque assays in MDCK cells. All experiments with ferrets were performed in accordance with the University of Tokyo’s Regulations for Animal Care and Use and approved by the Animal Experiment Committee of the Institute of Medical Science, the University of Tokyo.
Ferret transmission study
Pairs of ferrets were individually housed in adjacent wireframe cages that prevented direct and indirect contact between animals but allowed spread of influenza virus by respiratory droplets. Under anaesthesia, three ferrets per group were intranasally inoculated with 0.5 ml of 106 PFU/ml of Anhui/1, Dk/GM466, or CA04 (inoculated ferrets). One day after infection, three naïve ferrets (contact ferrets) were each placed in a cage adjacent to an ‘inoculated ferret’ (in these cages, infected and contact ferrets are separated by ~ 5 cm). Body weight and temperature were monitored every other day. Nasal washes were collected from ‘inoculated ferrets’ on day 1 after inoculation and from ‘contact ferrets’ on day 1 after co-housing, and then every other day (for up to 9 days) for virological examinations. The virus titres in nasal washes were determined by plaque assays in MDCK cells.
Experimental infection of cynomolgus macaques
Approximately two-year-old cynomolgus macaques (Macaca fascicularis) from Cambodia (obtained from Japan Laboratory Animals, Inc., Tokyo, Japan), weighing 2.2–2.8 kg and serologically negative by AniGen AIV antibody ELISA, which detects infection of all influenza A virus subtypes (Animal Genetics Inc., Tallahassee, FL) and neutralization against A/Osaka/1365/09 (pdm H1N1 2009), A/Kawasaki/UTK-4/09 (seasonal H1N1), A/Kawasaki/UTK-20/08 (H3N2), B/Tokyo/UT-E2/08 (type B), and A/duck/Hong Kong/301/78 (H7N2) viruses, were used in this study. Under anaesthesia, six and four macaques were inoculated with Anhui/1 or Dk/GM466 (107 PFU/ml each), respectively, through a combination of intratracheal (4.5 ml), intranasal (0.5 ml per nostril), ocular (0.1 ml per eye) and oral (1 ml) routes (resulting in a total infectious dose of 6.7 ×107 PFU). Body temperature was monitored at 0, 1, 3, 5, and 6 dpi by rectal thermometer. Nasal and tracheal swabs were collected at 1, 3, 5, and 6 dpi for virological examinations. Three Anhui/1- and two Dk/GM466-infected macaques per group were euthanized at 3 and 6 dpi for virological and pathological examinations. Virus titres were determined by plaque assays in MDCK cells. All experiments with macaques were performed in accordance with the Regulation on Animal Experimentation Guidelines at Kyoto University (February 5, 2007) and were approved by the Committee for Experimental Use of Nonhuman Primates in the Institute for Virus Research, Kyoto University.
Experimental infection of miniature pigs
Two- to three-month-old female specific-pathogen-free miniature pigs (NIBS line; Nippon Institute for Biological Science, Ome, Japan), which were serologically negative by neutralization assay for currently circulating human and swine influenza viruses, were used in this study. Baseline body temperatures were measured before infection. Four and two pigs were intranasally inoculated with 107 PFU (1 ml) of Anhui/1 or Dk/GM466, respectively. Body temperature was monitored daily. Nasal swabs were collected every day for virological examinations. Two pigs per group were euthanized at 3 dpi for virological and pathological examinations; the remaining two Anhui/1-inoculated pigs were euthanized at 6 dpi. Virus titres were determined by plaque assays in MDCK cells. All experiments with miniature pigs were performed in accordance with guidelines established by the Animal Experiment Committee of the Graduate School of Veterinary Medicine, Hokkaido University (Sapporo, Japan) and were approved by the Institutional Animal Care and Use Committee of Hokkaido University.
Experimental infection of chickens
Four-week-old female specific-pathogen-free chickens (Nisseiken Co. Ltd., Tokyo, Japan) were used in this study. Six chickens per group were intranasally inoculated with 2 ×106 PFU (0.2 ml) of Anhui/1 or Dk/GM466. Tracheal and cloacal swabs were collected every day for virological examinations. Three chickens per group were euthanized at 3 and 6 dpi for virological examinations. The virus titres in various organs and swabs were determined by plaque assays in MDCK cells. All experiments with chickens were performed in accordance with the National Institute of Infectious Diseases’ Regulations for Animal Care and Use and were approved by the Animal Experiment Committee of the National Institute of Infectious Diseases.
Experimental infection of quails
Four-month-old female quails (Yamanaka Koucho En, Kurashiki, Japan) were used in this study. Six quails per group were intranasally inoculated with 2 ×106 PFU (0.2 ml) of Anhui/1 or Dk/GM466. Tracheal and cloacal swabs were collected every day for virological examinations. Three quails per group were euthanized at 3 and 6 dpi for virological examinations. The virus titres in various organs and swabs were determined by plaque assays in MDCK cells. All experiments with quails were performed in accordance with the University of Tokyo’s Regulations for Animal Care and Use and were approved by the Animal Experiment Committee of the Institute of Medical Science, the University of Tokyo.
Pathological examination
Excised tissues of animal organs preserved in 10% phosphate-buffered formalin were processed for paraffin embedding and cut into 3-μm-thick sections. One section from each tissue sample was stained using a standard haematoxylin-and-eosin procedure, whereas another one was processed for immunohistological staining with a rabbit polyclonal antibody for type A influenza nucleoprotein (NP) antigen (prepared in our laboratory) that reacts comparably with all of the viruses tested in this study. Specific antigen–antibody reactions were visualized with 3,3′-diaminobenzidine tetrahydrochloride staining by using the DAKO LSAB2 system (DAKO Cytomation, Copenhagen, Denmark).
Cytokine and chemokine measurement
Macaque lung homogenates and serum samples were processed with the MILLIPLEX MAP Non-human Primate Cytokine/Chemokine Panel–Premixed 23-Plex (Merck Millipore, Billerica, MA). Array analysis was performed by using the Bio-Plex Protein Array system (Bio- Rad Laboratories).
Neuraminidase inhibition assay
In vitro NA activity of viruses was determined by using the commercially available NA- Fluor Influenza Neuraminidase Assay Kit (Applied Biosystems, Foster City, CA). Briefly, diluted viruses were mixed with the indicated amounts of oseltamivir, zanamivir, laninamivir, or peramivir and incubated at 37°C for 30 min. Methylumbelliferyl-N-acetylneuraminic acid (MUNANA) was then added as a fluorescent substrate, and the mixture was incubated at 37°C for 1 h. The reaction was stopped by adding 0.12 M Na2CO3 in 40% ethanol. The fluorescence of the solution was measured at an excitation wavelength of 355 nm and an emission wavelength of 460 nm, and the IC50 values were calculated.
Polymerase inhibitor sensitivity assay
8 ×105 MDCK cells were infected with approximately 50 PFU of viruses. After incubation at 37°C for 1 h, the viral inoculum was replaced with agarose medium containing various concentrations of favipiravir. After the cells were incubated at 37°C for 2 days, plaques were visualized by crystal violet staining and counted.
Antiviral sensitivity of viruses in mice
Under anaesthesia, six mice per group were intranasally inoculated with 103 or 104 PFU (50 μl) of Anhui/1, CA04, or a recombinant virus possessing the Shanghai/1 NA gene encoding NA-292K and the remaining genes from Anhui/1. At 2 h after inoculation, mice were treated with the following antiviral compounds: (1) oseltamivir phosphate: 4 or 40 mg per kg per 200 μl, administered orally twice a day for 5 days; (2) zanamivir: 0.8 or 8 mg per kg per 50 μl, administered intranasally once daily for 5 days; (3) laninamivir: 0.75 mg per kg per 50 μl, administered intranasally once during the entire experimental course; (4) favipiravir: 30 or 150 mg per kg per 200 μl, administered orally twice a day for 5 days; (5) or PBS intranasally (50 μl) and distilled water orally administered. For virological examinations, three mice per group were euthanized at 3 and 6 dpi. The virus titres in lungs were determined by plaque assays in MDCK cells.
Antigenicity characterization by HI assays
Anti-H7 HA monoclonal antibodies 46/6, 46/2, and 55/3 against A/seal/Massachusetts/1/80 (H7N7) virus were kindly provided by Dr. Robert G. Webster. The goat polyclonal antibody NR-9226 [raised against A/Netherlands/219/03 (H7N7)] was obtained from BEI Resources. The remaining antibodies, that is, mouse monoclonal antibodies B1275m and B1275m [raised against A/Netherlands/219/2003 (H7N7), MyBioSource, Inc.], 127-10023 [raised against A/FPV/Rostock/34 (H7N1), RayBiotech, Inc.], 10H9, 9A9, and 1H11 [raised against A/FPV/Rostock/34 (H7N1), HyTest Ltd.], and rabbit polyclonal antibody MBS432028 [raised against A/chicken/MD/MINHMA/2004 (H7N2), MyBioSource, Inc.], were commercially available. Anhui/1 propagated in embryonated chicken eggs or in MDCK cells was used in this study. Antibodies were serially diluted 2-fold with PBS in 96-well U-bottom microtitre plates and mixed with the amount of virus equivalent to 8 haemagglutination units, followed by incubation at room temperature for 30 min. After adding 50 μl of 0.5% chicken red blood cells, the mixtures were gently mixed and incubated at room temperature for a further 45 min. HI titres are expressed as the inverse of the highest antibody dilution that inhibited haemagglutination (Table S18). These data were used to select antibodies for glycan arrays.
Serology with human sera
Human sera, collected in Japan in November 2012 from 200 donors ranging in age from 20 to 63 years, were treated with receptor-destroying enzyme (Denka Seiken Co. Ltd., Tokyo, Japan). Two-fold serial dilutions of the treated sera were mixed with 100 PFU of Anhui/1 and incubated at 37°C for 1 h. MDCK cells were inoculated with the virus-serum mixtures and cultured for 3 days. The neutralizing activity of the sera was determined based on the cytopathic effects in inoculated cells. All experiments with human sera were approved by the Research Ethics Review Committee of the Institute of Medical Science, the University of Tokyo (approval number: 21-38-1117).
A total of 300 serum samples were also obtained from the serum bank of the National Institute of Infectious Diseases in Japan. Samples were collected from different regions of Japan during 2010–2011. Subjects were divided into 10 age groups, 30 samples per group, and analyzed for antibodies against Anhui/1 by use of the HI assay with 0.5% turkey red blood cells.
Glycan arrays
Glycan array analysis was performed on a glass slide microarray containing 6 replicates of 57 diverse sialic acid-containing glycans including terminal sequences and intact N-linked and O-linked glycans found on mammalian and avian glycoproteins and glycolipids34. Beta- propiolactone-inactivated viruses containing A(H7N9) virus HA and NA genes in the background of A/PR/8/34 (H1N1) virus were applied to the array at dilutions of 128–512 haemagglutination units per ml. After incubation at room temperature for 1 h, slides were washed and overlaid with a 1:200 dilution of rabbit or goat anti-H7 antibody for 1 h (selected based on antibody characterization with several H7 viruses; see Table S18), and finally with anti- rabbit IgG AlexaFluor-488 or anti-goat IgG AlexaFluor-647 (Invitrogen) at 10 μg/ml. Slides were then washed and scanned on a ProScanArray Express HT (PerkinElmer) confocal slide scanner to detect bound virus. A complete list of glycans present on the array is provided in Supplementary Materials (Table S9).
Electron microscopy
Anhui/1 was inoculated into 10-day-old embryonated chicken eggs and the allantoic membranes were collected 24 h after inoculation. They were then processed for ultrathin section electron microscopy and scanning electron microscopy as described previously9,35.
Statistical analysis
All statistical analyses were performed using JMP Pro 9.0.2 (SAS Institute Inc.). Statistically significant differences between the virus titres of Dk/GM466-infected mice and those of other mice were determined by using Welch’s t-test with Bonferroni’s correction. Comparisons of virus titres in antiviral sensitivity assays in mice were also done using Welch’s t-test or Student’s t-test on the result of the F-test. The resulting p-values were corrected by using Holm’s method.
Biosafety and biosecurity
All recombinant DNA protocols were approved by the University of Wisconsin-Madison’s Institutional Biosafety Committee after risk assessments were conducted by the Office of Biological Safety, and by the University of Tokyo’s Subcommittee on Living Modified Organisms, and, when required, by the competent minister of Japan. In addition, the University of Wisconsin-Madison Biosecurity Task Force regularly reviews the research program and ongoing activities of the laboratory. The task force has a diverse skill set and provides support in the areas of biosafety, facilities, compliance, security and health. Members of the Biosecurity Task Force are in frequent contact with the principal investigator and laboratory personnel to provide oversight and assure biosecurity. All experiments with H7N9 viruses were performed in enhanced BSL3 containment laboratories. Ferret transmission studies were conducted by two scientists with DVM and/or PhD degrees that each had more than 5 years of experience working with highly pathogenic influenza viruses and performing animal studies with such viruses. Our staff who work with ferrets, nonhuman primates, pigs, chickens, and quails wear disposable overalls and powered air-purifying respirators that filter the air. Biosecurity monitoring of the facility is ongoing. All personnel complete rigorous biosafety, BSL3, and Select Agent (for the US lab) training before participating in BSL3-level experiments. The principal investigator participates in training sessions and emphasizes compliance to maintain safe operations and a responsible research environment. The laboratory occupational health plan is in compliance with the University of Wisconsin-Madison and the University of Tokyo Occupational Health Programs.
Supplementary Material
Acknowledgments
We thank Dr. Yuelong Shu (Director of the WHO Collaborating Center for Reference and Research on Influenza, Director of the Chinese National Influenza Center, Deputy Director of the National Institute for Viral Disease Control and Prevention China CDC, Beijing, P.R. China) for A/Anhui/1/2013 (H7N9) and A/Shanghai/1/2013 (H7N9) viruses. We thank the IMSUT serum bank for providing human sera. We thank Dr. Robert Webster, St. Jude Children’s Research Hospital, Memphis, TN, for providing monoclonal antibody to A/seal/Massachusetts/1/80 (H7N7). Polyclonal Anti-Influenza Virus H7 Hemagglutinin (HA), A/Netherlands/219/03 (H7N7) (anti-serum, Goat), NR-9226 was obtained through the NIH Biodefense and Emerging Infections Research Resources Repository, NIAID, NIH. We thank Susan Watson for editing the manuscript, Drs. Tadaki Suzuki, Kenta Takahashi, Seiichiro Fujisaki and Hong Xu (all NIID, Japan) for helpful discussions, and Yuko Sato, Hiromi Sugawara, Aya Sato and Miho Ejima (all NIID, Japan) and Dr. Tomoyuki Miura (Kyoto University, Japan) for technical assistance. We thank Toyama Chemical Co., Ltd, for providing favipiravir, Daiichi Sankyo Co. Ltd. for providing laninamivir, F. Hoffmann-La Roche Ltd. for providing oseltamivir carboxylate, GlaxoSmithKline for providing zanamivir, and Shionogi & Co. Ltd. for providing peramivir. This work was supported by the Japan Initiative for Global Research Network on Infectious Diseases from the Ministry of Education, Culture, Sports, Science and Technology, Japan; by grants-in-aid from the Ministry of Health, Labour and Welfare, Japan; by ERATO (Japan Science and Technology Agency); by National Institute of Allergy and Infectious Diseases Public Health Service research grants AI099274 and AI058113 to JCP, and by an NIAID-funded Center for Research on Influenza Pathogenesis (CRIP, HHSN266200700010C) to YK.
Footnotes
Author contributions
T.W., M.K., S.F., M.I., S.Yamada, S.M., S.Yamayoshi, K.I-H., Y.S., E.T., M.H., S.W., E.A.M., G.N., H.K., T.O., J.C.P., M.T., and Y.K. designed the study; T.W., M.K., S.F., N.N., M.I., S.Y., S.M., S.Y., K.I-H., Y.S., E.T., R.M., T.N., M.H., H.I., D.Z., N.K., M.S., R.P.V., S.S., M.Okamatsu., T.T., Y.Y., N.F., K.G., H.K., I.I., M.I., Y.S-T., Y.S., R.U., R.Y., A.J.E., G.Z., S.F., J.P., A.H., Y.U., T.S., and H.H. performed the experiments; T.W., M.K., S.F., N.N., M.I., S.Y., S.M., S.Y., K.I-H., Y.S., E.T., R.M., T.N., M.H., H.I., D.Z., R.P.V., S.S., T.T., Y.T., H.K., E. K., and H. H. analyzed the data; T.W., S.F., N.N., E.T., R.M., M.H., R.P.V., M.Ozawa., G.N., T.O., J.C.P, H.H., M.T., and Y.K. wrote the manuscript; and Y.K. oversaw the project. T.W., M.K., S.F., N.N., and M.I. contributed equally to this work.
The authors declare the following competing financial interests: Y.K. has received speaker’s honoraria from Chugai Pharmaceuticals, Novartis, Daiichi-Sankyo Pharmaceutical, Toyama Chemical, Wyeth, GlaxoSmithKline and Astellas Inc.; grant support from Chugai Pharmaceuticals, Daiichi Sankyo Pharmaceutical, Toyama Chemical, Otsuka Pharmaceutical Co., Ltd; is a consultant for Crucell; and is a founder of FluGen. G.N. is a founder of FluGen.
References
- 1.2013 No authors listed( http://www.who.int/influenza/human_animal_interface/influenza_h7n9/ChinaH7N9JointMissionReport2013.pdf.
- 2.Gao R, et al. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med. 2013;368:1888–1897. doi: 10.1056/NEJMoa1304459. [DOI] [PubMed] [Google Scholar]
- 3.Kageyama T, et al. Genetic analysis of novel avian A(H7N9) influenza viruses isolated from patients in China, February to April 2013. Euro Surveill. 2013;18 [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Q, et al. Genomic signature and protein sequence analysis of a novel influenza A (H7N9) virus that causes an outbreak in humans in China. Microbes Infect. 2013 doi: 10.1016/j.micinf.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 5.Rogers GN, et al. Single amino acid substitutions in influenza haemagglutinin change receptor binding specificity. Nature. 1983;304:76–78. doi: 10.1038/304076a0. [DOI] [PubMed] [Google Scholar]
- 6.Hatta M, Gao P, Halfmann P, Kawaoka Y. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science. 2001;293:1840–1842. doi: 10.1126/science.1062882293/5536/1840. [pii] [DOI] [PubMed] [Google Scholar]
- 7.Subbarao EK, Kawaoka Y, Murphy BR. Rescue of an influenza A virus wild-type PB2 gene and a mutant derivative bearing a site-specific temperature-sensitive and attenuating mutation. J Virol. 1993;67:7223–7228. doi: 10.1128/jvi.67.12.7223-7228.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Z, et al. Molecular basis of replication of duck H5N1 influenza viruses in a mammalian mouse model. J Virol. 2005;79:12058–12064. doi: 10.1128/JVI.79.18.12058-12064.2005. 79/18/12058 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Itoh Y, et al. In vitro and in vivo characterization of new swine-origin H1N1 influenza viruses. Nature. 2009;460:1021–1025. doi: 10.1038/nature08260. nature08260 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van den Brand JM, et al. Severity of pneumonia due to new H1N1 influenza virus in ferrets is intermediate between that due to seasonal H1N1 virus and highly pathogenic avian influenza H5N1 virus. J Infect Dis. 2010;201:993–999. doi: 10.1086/651132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Min JY, et al. Classical swine H1N1 influenza viruses confer cross protection from swine-origin 2009 pandemic H1N1 influenza virus infection in mice and ferrets. Virology. 2010;408:128–133. doi: 10.1016/j.virol.2010.09.009. S0042-6822(10)00584-2 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maines TR, et al. Transmission and pathogenesis of swine-origin 2009 A(H1N1) influenza viruses in ferrets and mice. Science. 2009;325:484–487. doi: 10.1126/science.1177238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Munster VJ, et al. Pathogenesis and transmission of swine-origin 2009 A(H1N1) influenza virus in ferrets. Science. 2009;325:481–483. doi: 10.1126/science.1177127. 1177127 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Imai M, et al. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature. 2012;486:420–428. doi: 10.1038/nature10831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jackson S, et al. Reassortment between avian H5N1 and human H3N2 influenza viruses in ferrets: a public health risk assessment. J Virol. 2009;83:8131–8140. doi: 10.1128/JVI.00534-09. JVI.00534-09 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maines TR, et al. Lack of transmission of H5N1 avian-human reassortant influenza viruses in a ferret model. Proc Natl Acad Sci U S A. 2006;103:12121–12126. doi: 10.1073/pnas.0605134103. 0605134103 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herfst S, et al. Airborne transmission of influenza A/H5N1 virus between ferrets. Science. 2012;336:1534–1541. doi: 10.1126/science.1213362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang H, Carney P, Stevens J. Structure and Receptor binding properties of a pandemic H1N1 virus hemagglutinin. PLoS Curr. 2010;2:RRN1152. doi: 10.1371/currents.RRN1152. k/-/-/250q41vwql45q/3 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamada S, et al. Haemagglutinin mutations responsible for the binding of H5N1 influenza A viruses to human-type receptors. Nature. 2006;444:378–382. doi: 10.1038/nature05264. nature05264 [pii] [DOI] [PubMed] [Google Scholar]
- 20.Chutinimitkul S, et al. In vitro assessment of attachment pattern and replication efficiency of H5N1 influenza A viruses with altered receptor specificity. J Virol. 2010;84:6825–6833. doi: 10.1128/JVI.02737-09. JVI.02737-09 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Srinivasan K, Raman R, Jayaraman A, Viswanathan K, Sasisekharan R. Quantitative Characterization of Glycan-Receptor Binding of H9N2 Influenza A Virus Hemagglutinin. PLoS One. 2013;8:e59550. doi: 10.1371/journal.pone.0059550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chandrasekaran A, et al. Glycan topology determines human adaptation of avian H5N1 virus hemagglutinin. Nat Biotechnol. 2008;26:107–113. doi: 10.1038/nbt1375. nbt1375 [pii] [DOI] [PubMed] [Google Scholar]
- 23.Belser JA, et al. Contemporary North American influenza H7 viruses possess human receptor specificity: Implications for virus transmissibility. Proc Natl Acad Sci U S A. 2008;105:7558–7563. doi: 10.1073/pnas.0801259105. 0801259105 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKimm-Breschkin JL, et al. Mutations in a conserved residue in the influenza virus neuraminidase active site decreases sensitivity to Neu5Ac2en-derived inhibitors. J Virol. 1998;72:2456–2462. doi: 10.1128/jvi.72.3.2456-2462.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Centers for Disease C & Prevention. Emergence of Avian Influenza A(H7N9) Virus Causing Severe Human Illness - China, February-April 2013. MMWR Morb Mortal Wkly Rep. 2013;62:366–371. [PMC free article] [PubMed] [Google Scholar]
- 26.Wetherall NT, et al. Evaluation of neuraminidase enzyme assays using different substrates to measure susceptibility of influenza virus clinical isolates to neuraminidase inhibitors: report of the neuraminidase inhibitor susceptibility network. J Clin Microbiol. 2003;41:742–750. doi: 10.1128/JCM.41.2.742-750.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu X, et al. Poor responses to oseltamivir treatment in a patient with influenza A (H7N9) virus infection. Emerg Microbes & Infections. 2013;2:e27. doi: 10.1038/emi.2013.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jakiela B, Brockman-Schneider R, Amineva S, Lee WM, Gern JE. Basal cells of differentiated bronchial epithelium are more susceptible to rhinovirus infection. Am J Respir Cell Mol Biol. 2008;38:517–523. doi: 10.1165/rcmb.2007-0050OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neumann G, et al. Generation of influenza A viruses entirely from cloned cDNAs. Proc Natl Acad Sci U S A. 1999;96:9345–9350. doi: 10.1073/pnas.96.16.9345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 31.Makarova NV, Ozaki H, Kida H, Webster RG, Perez DR. Replication and transmission of influenza viruses in Japanese quail. Virology. 2003;310:8–15. doi: 10.1016/s0042-6822(03)00094-1. S0042682203000941 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu J, et al. Highly pathogenic H5N1 influenza virus infection in migratory birds. Science. 2005;309:1206. doi: 10.1126/science.1115273. 1115273 [pii] [DOI] [PubMed] [Google Scholar]
- 33.Zhu H, et al. Infectivity, Transmission, and Pathology of Human H7N9 Influenza in Ferrets and Pigs. Science. 2013 doi: 10.1126/science.1239844. science.1239844 [pii] [DOI] [PubMed] [Google Scholar]
- 34.Xu R, et al. A recurring motif for antibody recognition of the receptor-binding site of influenza hemagglutinin. Nature structural & molecular biology. 2013;20:363–370. doi: 10.1038/nsmb.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Noda T, et al. Architecture of ribonucleoprotein complexes in influenza A virus particles. Nature. 2006;439:490–492. doi: 10.1038/nature04378. nature04378 [pii] [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.