Abstract
When access to molecular oxygen is restricted, Mycobacterium tuberculosis (Mtb) can respire an alternative electron acceptor, nitrate. We found that Mtb within infected primary human macrophages in vitro at physiologic tissue oxygen tensions respired nitrate, generating copious nitrite. A strain of Mtb lacking a functioning nitrate reductase was more susceptible than wild-type Mtb to treatment with isoniazid during infection of macrophages. Likewise, nitrate reductase-deficient Mtb was more susceptible to isoniazid than wild-type Mtb in axenic culture, and more resistant to hydrogen peroxide. These phenotypes were reversed by the addition of exogenous nitrite. Further investigation suggested that nitrite might inhibit the bacterial catalase. To the extent that Mtb itself is the most relevant source of nitrite acting within Mtb, these findings suggest that inhibitors of Mtb's nitrate transporter or nitrate reductase could enhance the efficacy of isoniazid.
Keywords: Hydrogen peroxide, isoniazid, Mycobacterium tuberculosis, nitrate reductase, nitrite
Introduction
Nitrate (NO3−) and nitrite (NO2−) arise in the mammalian host from three major sources – dietary, bacterial, and host. Dietary nitrate is absorbed into the blood. Some is reduced by oral and gastrointestinal commensals, producing nitrite, which is also absorbed. Nitric oxide synthases (NOSs), including those that are constitutively expressed, produce nitric oxide, which autoxidizes to nitrate and nitrite. Even in healthy individuals, in whom relatively little of the high-output NOS isoform (iNOS) is expressed (Nicholson et al. 1996), nitrate is present in plasma at levels of 20–40 μmol/L (Lundberg and Weitzberg 2010). In patients with tuberculosis (TB), iNOS and other NOSs are expressed in granulomas (Choi et al. 2002; Lundberg et al. 2009).
At oxygen tensions of 1% or lower, Mtb itself can produce nitrite as it maintains adenosine-5'-triphosphate (ATP) synthesis and redox homeostasis by reducing an alternative electron acceptor, nitrate. Mtb constitutively expresses the narGHJI operon, which encodes Mtb's only functional nitrate reductase (Wayne and Sohaskey 2001; Sohaskey and Wayne 2003). However, transport of nitrate into the bacterium depends on transcription of a nitrate transporter encoded by narK2, a member of the dormancy survival (DOS) regulon (Sherman et al. 2001; Voskuil et al. 2003; Giffin et al. 2012). The narK2 gene is expressed in axenically cultured Mtb only when oxygen levels fall below 1%. Induction of narK2 markedly enhances mycobacterial nitrate respiration (Sohaskey and Wayne 2003). RNA transcripts from narG, which encodes a subunit of the nitrate reductase, and narX, an inactive fused nitrate reductase downstream of narK2 in a shared operon, have been detected in lungs of patients with TB (Fenhalls et al. 2002; Sohaskey and Wayne 2003; Rachman et al. 2006), and RNA transcripts encoding the nitrate transporter, narK2, were elevated in the lungs of Mtb-infected mice (Shi et al. 2005). Thus, mycobacterial nitrate reduction likely occurs in the infected host.
Our laboratory recently converted our culture system for primary human macrophages from conventional gas phase conditions of 21% O2, 5% CO2 to 10% O2, 5% CO2 on the grounds that 10% O2 is physiologic for tissue macrophages (Vogt and Nathan 2011). To our surprise, infection of the macrophages in vitro with wild-type Mtb under 10% oxygen resulted in extensive accumulation of nitrite in the supernatant. Evidence will be presented elsewhere that macrophages were not the source of nitrite in this system. In contrast, as shown below, Mtb's NarG was the source, despite the nonhypoxic level of oxygen in the gas phase.
Wayne has proposed that nitrate respiration supports the establishment of a nonreplicating persistent state (NRP) in response to hypoxia (Wayne and Hayes 1998). We hypothesized that Mtb lacking the capacity for nitrate respiration might demonstrate an impaired entrance into NRP and therefore remain more susceptible to drugs that target replicating Mtb. The goal of the present study was to assess the contribution of nitrate respiration to Mtb's sensitivity to the drugs isoniazid (INH), ethambutol, streptomycin, and rifampicin. Compared to wild-type Mtb, narG-deficient Mtb, which is unable to produce nitrite, was selectively hypersusceptible to INH, as well as hyperresistant to hydrogen peroxide. INH requires oxidative activation by the mycobacterial catalase/peroxidase KatG in order to become cidal. The resultant isonicotinoyl radical adducts with cellular pyridine nucleotides and potently inhibits InhA, a member of type II fatty acid synthesis pathway involved in cell wall mycolic acid biosynthesis (Winder and Collins 1970; Rozwarski 1998). These findings imply that the action of INH on Mtb may be blunted by nitrite contributed by the host, the pathogen, or both.
Methods
Isolation and differentiation of primary human monocytes
Isolation of human monocytes and their differentiation into macrophages was as described (Vogt and Nathan 2011). In brief, heparinized peripheral blood was collected by venipuncture from healthy human donors who provided informed consent under an IRB approved protocol. Peripheral blood mononuclear cells were first isolated by centrifugation of whole blood over Ficoll-Paque (GE Healthcare, Uppsala, Sweden). The buffy coats were collected and monocytes were isolated by positive selection using magnetic beads conjugated to anti-CD14 antibodies (Miltenyi Biotec, Auburn, CA). Following isolation, the human monocytes were plated at a density of 500,000 cells/mL per well of a 96 well plate. The culture medium consisted of 60% Roswell Park Memorial Institute medium (RPMI) 1640, supplemented with 1% glutamax, 40% human plasma and granulocyte-macrophage colony-stimulating factor (GM-CSF) and tumor necrosis factor alpha (TNFα) (0.5 ng/mL each). Thirty percent of the total culture volume was replaced with fresh medium and cytokines every 3–4 days. Replacing fresh medium was conducted as rapidly as possible in room air before the cells were placed back in the low oxygen incubator. Differentiation of the monocytes was conducted over 2 weeks in our standard medium at 10% O2 and 5% CO2 at 37°C in a humidified atmosphere in a chamber flushed with N2 under the control of a PRoOX sensor and ProCO2 regulator (BioSpherix, Lacona, NY). They were then activated with interferon gamma (IFNγ) (5 ng/mL) before infection with Mycobacterium tuberculosis (Mtb) the following day at the desired multiplicity of infection (MOI).
Preparation of Mtb and infection of human macrophages with Mtb
Mycobacterium tuberculosis H37Rv was grown in Middlebrook 7H9 broth supplemented with 0.2% glycerol, 0.5% bovine serum albumin (BSA), 0.2% dextrose, and 0.085% NaCl with 0.05% Tween 80. The narG-deficient and complemented strains were generated as described (Stermann et al. 2003). Cultures were started with 1 mL stock originally frozen at −80°C in log phase and then grown over 4–5 days to optical densities (OD) of 0.5–1.25 before the start of an experiment. A single-cell suspension was generated by centrifugation at 120 g for 10 min. For macrophages infection, roughly 200 × 106 bacteria were centrifuged, in order to be able to observe a pellet. The 7H9 was completely removed and the cells were washed with phosphate buffered saline (PBS). The bacteria were then resuspended in culture medium and the desired number of Mtb was added to the macrophage culture.
Certain experiments required that we remove the nitrate-containing medium from the cell culture. In this case, 1 day prior to infection the cells were washed with room temperature PBS three times and a low-nitrate formulation of Dulbecco's Modified Eagle Medium (DMEM) with 10% human plasma was added along with 0.5 ng/mL GMCSF and TNFα and 5 ng/mL IFNγ.
Measurement of nitrite
Nitrite levels produced in the coculture supernatants of infected macrophages were measured by the Griess assay. Briefly, 100 μL of supernatant was removed and to this were added 50 μL of 2% sulfanilamide with 5% phosphoric acid and 50 μL of 0.2% N-1-napthylethylenediamine dihydrochloride. Nitrite standards were prepared in the same medium used to culture the macrophages. Absorbance was measured at 550 nm immediately after addition of the reagents to the supernatant samples.
Treatment of infected macrophages
Macrophages were incubated with Mtb at the indicated MOI for 4–5 h under 10% oxygen. The supernatant was removed and the macrophages were washed three times with room temperature PBS to remove extracellular bacteria. Fresh medium was added to the cells, followed by the addition of nitrite, nitrate, and/or an antimycobacterial drug such as INH. The cultures were then returned to the low oxygen incubator and incubated for a subsequent 3 days with the desired compound. Drugs were freshly dissolved before each experiment in dimethyl sulfoxide (DMSO), except for streptomycin, which was dissolved in water.
Colony forming units determination from infected macrophages
Following incubation with Mtb for 3 days, the macrophages were inspected by microscopy to ensure continued confluence and adherence to the well. The supernatant was collected and the cells were gently washed three times with room temperature PBS to wash away remaining compounds. Following the wash steps, the cells were again visualized to confirm that the monolayer was intact and then lysed by incubation in 0.5% Triton X-100 (Sigma-Aldrich, St. Louis, MO) for 10 min at 37°C. The lysate was serially diluted in 0.1% Triton X-100 and plated on Middlebrook 7H11 agar with 10% oleic acid–albumin–dextrose–catalase (OADC) enrichment (Becton Dickinson, Franklin Lakes, NJ) supplemented with 0.5% glycerol. Colonies were enumerated following 3 weeks of culture at 37°C, in 21% O2, 5% CO2.
Treatment of Mtb in axenic culture
Mycobacterium tuberculosis was added at the indicated OD to 96 well plates in 7H9 broth with or without 5 mmol/L nitrate or the indicated concentration of nitrite, supplemented with 10 ADNaCl (0.2% glycerol, 0.5% BSA, 0.2% dextrose, 0.085% NaCl) and 0.05% Tween 80. The indicated antibiotic was added immediately and the plate was incubated in 1% oxygen for 3 days or at 21% oxygen for 5 days, as indicated. The Mtb was resuspended, serially diluted in 0.1% Triton-X 100 (Sigma-Aldrich), and plated for colony forming units (CFU) as above. Alternatively, for OD measurements, the Mtb was resuspended in the culture medium and the absorbance was read at 580 nm on the fifth day of incubation with the indicated compound. In the case of exposure to hydrogen peroxide, the plate was incubated in 1% oxygen for 8 h with or without 1 mmol/L nitrate or the indicated concentration of nitrite. At this point, the indicated concentration of hydrogen peroxide was added and the cells were incubated overnight at 1% oxygen before plating for CFU.
KatG western blot and measurement of KatG dependent oxidation of INH
A minimum of 4 × 109 bacteria per condition were suspended in 7H9, 10% ADNaCl at an OD of 0.2 in the presence or absence of 2.5 mmol/L nitrite. Cultures were incubated for 1 day in 1% oxygen. The cultures were collected by centrifugation and resuspended in 200 μL of lysis buffer containing 50 mmol/L NaPO4, 1 mM phenylmethylsulfonyl fluoride (PMSF), and 4× complete protease inhibitor cocktail (Roche, Basel, Switzerland). The cells were lysed in a bead-beating homogenizer with silica beads three times with intermittent incubation on ice. The lysate was collected and filter sterilized and concentrated at 4°C to ∼100 μL using Amicon Ultra-0.5 mL centrifugal filters from Millipore (Billerica, MA) with a molecular weight cutoff of 3 kDa. Protein concentration was measured using the Bio-Rad (Hercules, CA) assay and equal amounts of protein from each sample were loaded into 7.5% reducing gels. After electrophoresis, proteins were transferred onto a polyvinylidene difluoride membrane, blocked with Odyssey blocking buffer from LI-COR Biosciences (Lincoln, NE) and stained with anti-KatG antibody (obtained through BEI Resources, NIAID, NIH: Monoclonal Anti-Mycobacterium tuberculosis KatG, gene Rv1908c, clone IT-57 [CDA4] [culture supernatant], NR-13793) and anti-DlaT antibody (1:1000) and then visualized with fluorophore-coupled secondary antibodies (Venugopal et al. 2011). To test the degree of KatG-dependent oxidation of INH, 50 μg of total cell lysate was loaded into 7.5% nonreducing gels. After electrophoresis, the gel was incubated at room temperature for 15–30 min in 50 mL of NaPO4 buffer containing 62 mg INH, and 2 mmol/L nitroblue tetrazolium (NBT) at pH 7 to which hydrogen peroxide was added to a final concentration of 800 μmol/L (Saint-Joanis et al. 1999).
Quantitative polymerase chain reaction of RNA from Mtb growing in macrophages or broth culture
Five million human macrophages were cultured per T25 flask in 10% oxygen and infected with wild-type, narG-deficient, or the complemented strains of Mtb at an MOI of 40 for 10 h. Supernatant was removed and the monolayer was washed two times with room temperature PBS. Trizol (2 mL) was added to each flask and the cells were detached using a rubber policeman. For broth cultures, at least 2.5 × 109 bacteria per condition were incubated in 1% oxygen for 3 days with or without 2.5 mmol/L nitrite and 0.1 μg/mL INH. Then an equal volume of 5 mol/L GTC buffer containing 5M guanidine thiocyanate, 25 mmol/L sodium citrate, 20 mmol/L N-lauryl-sarcosine, 0.7% v/v β- mercaptoethanol was added. The bacteria were pelleted by centrifugation and 1 mL of Trizol was added per sample. The suspension was beaten with silica beads, three times. RNA was extracted using an RNeasy kit (Qiagen, Venlo, Limburg, Netherlands) in accordance with the manufacturer's instructions with the exception that off column DNAse digestion was performed for 2 h at 37°C. To generate cDNA, total RNA (900 ng) was reverse transcribed using the GeneAmp RNA polymerase chain reaction (PCR) Kit (Life Technologies, Carlsbad, CA). Quantitative RT-PCR was performed using gene-specific primers (Life Technologies) and the SuperScript III Platinum Two Step real-time quantitative PCR (qRT-PCR) Kit (Life Technologies) with a 7900HT Fast Real Time PCR System (Life Technologies). Each experiment contained experimental triplicates. All reported values were within the linear range of the primers and the experimental results were normalized to 16S rRNA values.
Statistical analysis
Statistical analysis was performed as indicated in the figure legends using the standard statistical software Prism version 5.0f for Macintosh, GraphPad Software, San Diego, CA.
Results
Nitrate respiration enhanced mycobacterial resistance to isoniazid within primary human macrophages
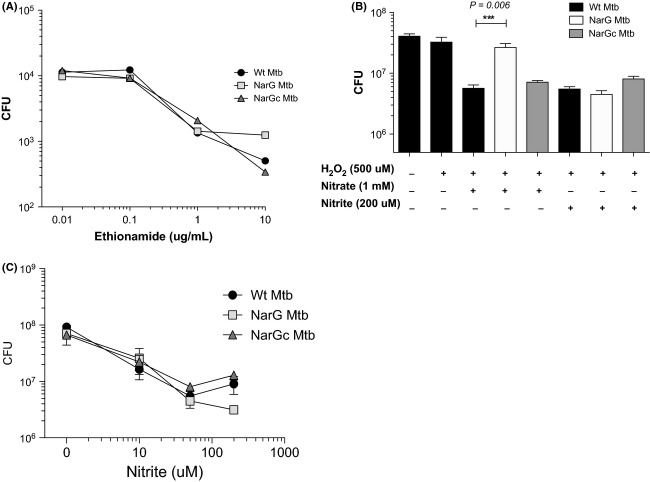
We observed no difference in survival of wild-type (Wt Mtb) and narG-deficient Mtb (NarG Mtb) during infection of primary human macrophages under 10% O2 and 5% CO2 (Fig. 1A). During the course of infection, large quantities of nitrite arose in the supernatant of macrophages infected with wild-type Mtb, which was not observed in the supernatants of macrophages infected with narG-deficient Mtb (Fig. 1B). To test the role of nitrate reduction on the susceptibility of Mtb to antibiotics, we infected macrophages with wild-type, narG-deficient, and complemented strains (NarGc Mtb) and treated the cultures with drugs. Mtb deficient in narG was hypersusceptible to INH, with a 10-fold decrease in the MBC99 compared to wild type (Fig. 1C–D). In contrast, narG made no observable difference to the antibacterial actions of streptomycin, rifampicin, or ethambutol on Mtb within macrophages (Fig. S1A–F).
Figure 1.
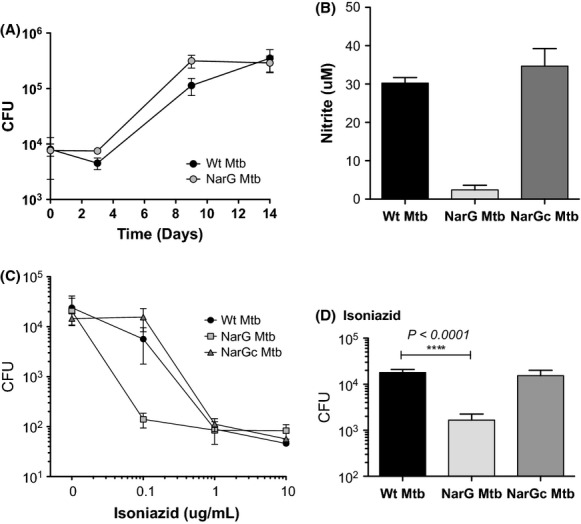
Effect of nitrate respiration on mycobacterial resistance to isoniazid within primary human macrophages. (A) Survival of wild-type and narG-deficient (NarG) Mtb within macrophages (MOI: 0.1, corresponding to 10,000 bacteria). (B) Nitrite levels arising in the supernatant of macrophages 3 days after infection with wild-type Mtb (Wt Mtb), NarG Mtb, and the complemented strain (NarGc Mtb) (MOI: 5). (C) Susceptibility of Mtb to INH within infected macrophages (MOI: 0.1) over 3 days. Means ± SEM from one experiment representative of three independent experiments using cells from different donors. (D) As in (C), pooling results from the three donors (means ± SEM) at a single INH concentration administered to the culture for 3 days (0.1 μg/mL). The p value was determined by an unpaired t-test.
Addition of nitrite reversed the hypersusceptibility of narG-deficient Mtb to isoniazid
The hypersusceptibility of narG-deficient Mtb to INH may have resulted from loss of the metabolic energy generated by nitrate reduction or from failure to produce nitrite. To distinguish between these possibilities, we added exogenous nitrite to the medium of infected macrophage cultures treated with INH. Because the standard culture medium consisted of 40% human plasma and 60% RPMI 1640, it contained high concentrations of nitrate. Therefore, to limit the accumulation of nitrite produced by wild-type Mtb, we replaced the medium immediately prior to infection with a low-nitrate medium (to minimize perturbation we refrained from washing the cells thoroughly; thus, medium exchange was not complete). Addition of exogenous nitrite enhanced bacterial survival in the presence of INH in a dose-dependent manner (Fig. 2A–C). That this effect was less striking in the case of wild-type Mtb was likely due to residual nitrate in the medium, allowing for low-level nitrite production (Fig. 2A).
Figure 2.
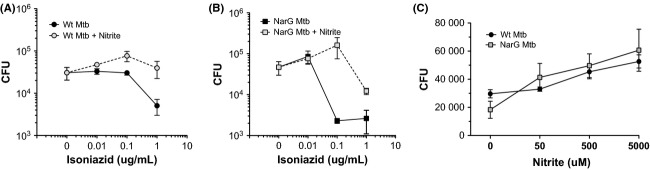
Impact of exogenous nitrite on the hypersusceptibility of narG-deficient Mtb to isoniazid. (A, B) The effect of nitrite on the survival of (A) wild-type or (B) narG-deficient (NarG) Mtb within INH-treated infected macrophages (MOI: 0.1). The standard culture medium was replaced with low-nitrate medium. Where indicated by the dashed lines, exogenous sodium nitrite (final concentration 1 mmol/L) was added to the culture medium along with the indicated concentrations of INH. CFU were harvested 3 days later. (C) Survival of wild-type and narG-deficient (NarG) Mtb within macrophages treated with increasing concentrations of nitrite in the presence of a single concentration of INH (0.1 μg/mL). The standard culture medium was replaced with low-nitrate medium and exogenous sodium nitrite was added at the indicated final concentrations. Each result is representative of at least two independent experiments (means ± SEM).
Mtb deficient in narG Mtb was hypersusceptible to isoniazid in axenic culture
Mycobacterium tuberculosis respires nitrate when infecting human macrophages under 10% oxygen, but in axenic culture, as noted, nitrate respiration is barely detectable until oxygen is lowered to 1% or less. This implied that within macrophage phagosomes, the functional level of oxygen perceived by Mtb was far lower than the level present in the gas phase. The nitrate reductase assay is routinely used in clinical laboratories to identify drug resistant Mtb (Angeby et al. 2002). Although the assay is carried out under room air (21% oxygen), nitrate reductase activity is measured at the end of a prolonged incubation period, which may indicate that the medium in which Mtb is incubated becomes hypoxic. Alternatively, while aerobic cultures of Mtb do not express the nitrate transporter, narK2, nitrate may gradually diffuse across the plasma membrane and be reduced to nitrite. In order to test the role of nitrate respiration in Mtb's susceptibility to INH in the absence of macrophages, we cultured wild-type and narG-deficient Mtb in 21% oxygen or 1% oxygen. No measurable survival differences were observed for the two strains in room air in response to isoniazid, streptomycin, or rifampicin (Fig. 3A–C). Under 1% oxygen, however, the survival of narG-deficient INH-treated Mtb dropped by 2–3 log10 as compared to wild-type and complemented strains (Fig. 3D), which was due to the induction of the nitrate reductase system. Next, we examined survival as a function of time rather than concentration. A single application of INH led to a drop of 2 log10 in CFU of the narG-deficient strain over 9 days, while the CFU of the wild-type and complemented strains remained constant (Fig. 3E). Exogenous nitrite enhanced the survival of the narG-deficient and wild-type strains cultured in the absence of nitrate and treated with INH (Fig. 3F).
Figure 3.
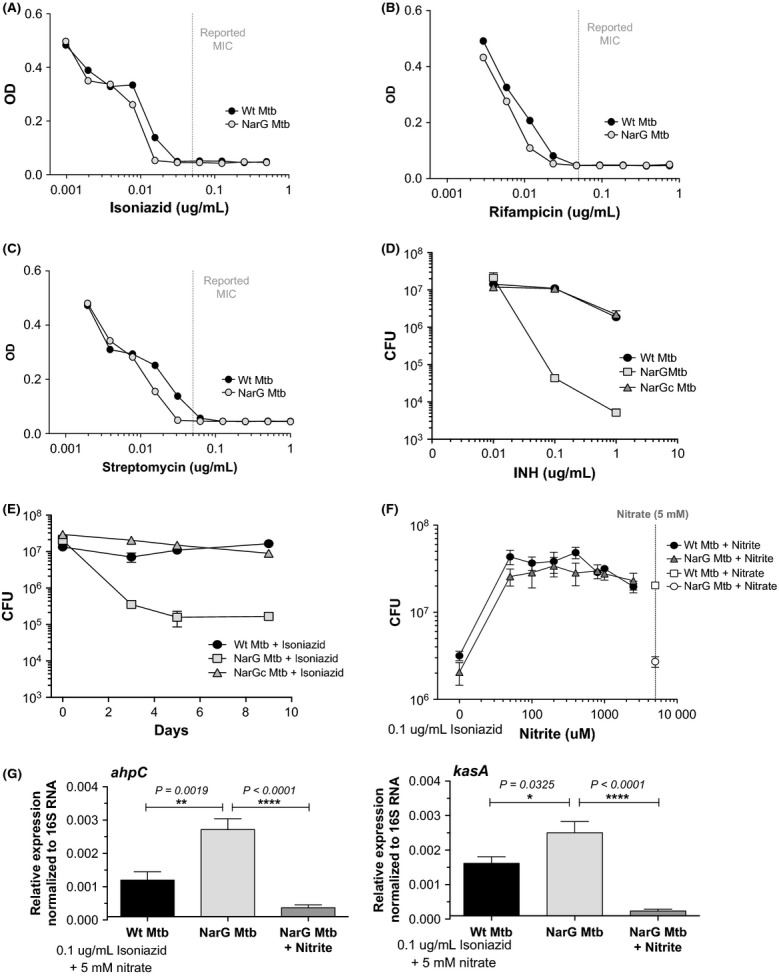
Impact of narG deficiency on susceptibility of Mtb to isoniazid in axenic culture. (A–C) Survival of wild-type and narG-deficient Mtb in the presence of the indicated concentrations of (A) isoniazid, (B) rifampicin, or (C) streptomycin in 21% oxygen over 5 days, initial (OD 0.1). Published MICs are provided for reference (Wanger and Mills 1996). (D, E) Survival of wild-type, narG-deficient (NarG), and complemented (NarGc) strains cultured in 1% oxygen (D) for 3 days with the indicated concentrations of INH or (E) as a function of time over 9 days with 0.1 μg/mL INH. (F) Survival of wild-type and narG-deficient Mtb incubated with increasing concentrations of exogenous nitrite or one fixed dose of nitrate, as indicated, over 3 days in 1% oxygen with 0.1 μg/mL INH. Results for (A–F) are means ± SEM representative of two independent experiments. (G) Expression of transcripts of ahpC and kasA by wild-type and narG-deficient Mtb treated with 0.1 μg/mL INH and 5 mmol/L nitrate for 16 h at 1% oxygen relative to 16S RNA. Where indicated, 2.5 mmol/L nitrite was also added. Bars indicate means ± SEM from three independent experiments analyzed by unpaired t-tests.
Following INH treatment and its subsequent activation by KatG, Mtb induces the expression of kasA and ahpC (Wilson et al. 1999). The narG-deficient strain demonstrated greater induction of kasA and ahpC RNA transcripts than the wild-type strain in response to 0.1 μg/mL INH, and induction was attenuated following treatment with nitrite (Fig. 3G). INH requires oxidative activation by the mycobacterial catalase/peroxidase KatG in order to become bioactive. These results suggest that when presented with the same concentration of INH under conditions that could sustain nitrate respiration by wild-type Mtb, narG-deficient Mtb converted a larger proportion of the INH prodrug to its active form than wild-type Mtb. This implied that the production of nitrite during respiration of nitrate by wild-type Mtb might partially inhibit KatG.
narG-deficient Mtb was not hypersusceptible to treatment with ethionamide
To further investigate the role of KatG in mediating the hypersusceptibility of narG-deficient Mtb to INH, we infected human macrophages with Mtb and treated the cultures with ethionamide (ETH). ETH and INH are close chemical analogs. Both are prodrugs that when activated inhibit the same target, InhA. However, ETH is activated by the monoxygenase EtaA rather than by KatG (Baulard et al. 2000; DeBarber et al. 2000). Survival of wild-type and narG-deficient Mtb treated with ETH within infected human macrophages did not differ (Fig. 4a). Therefore, the INH susceptibility of narG-deficient Mtb is unlikely to be due to differential dependence on InhA and may instead be due to differences in the activation of INH by KatG.
Figure 4.
Impact of narG deficiency on susceptibility of Mtb to ethionamide and hydrogen peroxide. (A) Survival of wild-type and narG-deficient Mtb within infected primary human macrophages (MOI: 0.1) treated with the indicated concentrations of ethionamide for 3 days at 10% oxygen. (B) Survival of Mtb in axenic culture treated with nitrate or nitrite for 8 h in 1% oxygen, to allow for nitrate respiration to occur, and then treated with hydrogen peroxide (500 μmol/L) and incubated in 1% oxygen overnight (initial OD580: 0.1). These are pooled data from two independent experiments and were analyzed by an unpaired t-test. (C) Survival of wild-type, narG-deficient (NarG Mtb), and complemented strains (NarGc Mtb) (initial OD580: 0.1) following treatment with both hydrogen peroxide (500 μmol/L) and increasing concentrations of nitrite as indicated. Individual experiments representative of at least two independent experiments. Bars indicate means ± SEM.
Nitrate reduction enhanced the lethality of hydrogen peroxide
Levels of KatG mRNA determined by qRT-PCR and KatG protein detected by western blot did not differ among wild-type, narG-deficient, and complemented strains of Mtb (Fig. S2B–C). Moreover, we were unable to measure differences in the extent of KatG-dependent oxidation of INH via NBT staining of polyacrylamide gel electrophoregrams of lysates obtained from the three strains (data not shown). Nonetheless, to help assess whether the catalytic activity of KatG might differ between wild-type and narG-deficient Mtb, the strains were incubated for several hours with nitrite or nitrate at 1% oxygen to allow for nitrate respiration and then treated with hydrogen peroxide. We reasoned that if the activity of KatG were attenuated by nitrite, the wild-type and complemented strains might be more susceptible to hydrogen peroxide than narG-deficient Mtb. In accord with this hypothesis, narG-deficient Mtb was more resistant to hydrogen peroxide than the other two strains (Fig. 4B). The enhanced mycobactericidal effect of hydrogen peroxide on the wild type and complemented strains was also observed following the addition of increasing concentrations of exogenous nitrite in the absence of nitrate (Fig. 4C and S2B).
Discussion
Herein, we report that deficiency of narG increases the susceptibility of Mtb to INH and decreases its susceptibility to hydrogen peroxide. Prior to this work, the knockout of only one gene, cydC, had been reported to increase the susceptibility of Mtb to INH (Dhar and McKinney 2010). The mechanism in the present study most likely involves nitrite-mediated impairment of the activity of KatG that takes place when wild-type Mtb reduces nitrate to nitrite, which is absent in narG-deficient Mtb. Although nitrate is usually omitted from mycobacterial culture media, it is a physiologic constituent of human body fluids. Although Mtb does not significantly reduce nitrate to nitrite in axenic culture unless the culture is hypoxic, we found that Mtb does reduce nitrate to nitrite when cultured within macrophages at physiologic oxygen tensions. Thus, phenotypic INH resistance arising from Mtb's nitrate respiration is likely to occur in the Mtb-infected host.
Interactions of catalase and other peroxidases with nitrite are complex (Klebanoff 1993; van der Vliet et al. 1997; Eiserich et al. 1998; Battistuzzi et al. 2010). When we incubated Mtb with nitrite, lysed the cells, separated the proteins by nonreducing polyacrylamide gel electrophoresis and incubated the gel with INH and a tetrazolium dye as described (Saint-Joanis et al. 1999), we did not see impairment in the activity of catalase compared to lysates from Mtb not exposed to nitrite (not shown). However, the biochemical environment within intact mycobacteria differs markedly from that of gels in room air, and tetrazolium is not a physiologic oxidant for INH. Moreover, katG mutations that cause INH resistance do not necessarily impact the rate of INH-NAD adduct formation by KatG relative to wild-type Mtb, pointing to the potential for dissociation between the functional effects of KatG mutations and the results of biochemical assays for KatG activity (Musser et al. 1996; Wengenack et al. 2004; Ghiladi et al. 2005; Cade et al. 2010).
The following evidence supports the hypothesis that nitrite interfered with the INH-activating function of catalase in the intact mycobacterium: Mtb deficient in narG was both more susceptible to INH and more resistant to hydrogen peroxide than wild-type Mtb. Second, narG-deficient Mtb was not hypersusceptible to treatment with ETH. Third, compared to wild-type and complemented strains, narG-deficient Mtb demonstrated higher expression of ahpC and kasA, two genes induced by treatment with INH, and their expression was reduced by the addition of nitrite to the cell culture. Taken together, these results suggest that Mtb lacking narG achieves greater KatG-mediated activation of INH than wild-type Mtb. Lastly, in agreement with this hypothesis, narG-deficient Mtb better resisted treatment with hydrogen peroxide. The mutation of genes other than katG can also cause INH resistance. For example, mutations that impair the catalytic activities of Ndh, a type II NADH dehydrogenase, and MshA and MshC, which contribute to mycothiol biosynthesis, can cause INH resistance. However, inactivating mutations in any one of these genes also cause ETH resistance (Vilchèze et al. 2005; Hazbon et al. 2006; Vilcheze et al. 2011a), which was not observed in this study.
Nitric oxide or nitrite at low pH, which generates nitric oxide (Lundberg et al. 2008), can potentiate the bactericidal activity of hydrogen peroxide toward Escherichia coli (Klebanoff 1993; Pacelli et al. 1995; Woodmansee and Imlay 2003; Lundberg and Weitzberg 2010). However, to our knowledge, this is the first report that nitrite at neutral pH can synergistically enhance the bactericidal action of hydrogen peroxide. Production of hydrogen peroxide by phagocytic cells is an important component of host antibacterial defense (Nathan and Shiloh 2000). Mycobacterial nitrate respiration may enhance the lethality of host reactive oxygen species in vivo. This may result from inhibition of KatG by nitrite or from inhibition of a variety of antioxidant defense proteins, including KatG, by products more reactive than nitrite to which nitrite can give rise. In E. coli, nitrate reductase is an important source of species reactive enough to nitrosylate proteins, as evidenced by an 80% reduction in protein S-nitrosylation in narG-deficient as compared to wild-type bacteria (Ralt et al. 1988; Corker 2003; Seth et al. 2012). Rhee et al. (2005) identified 29 mycobacterial proteins that were S-nitrosylated when Mtb was treated with nitrite at low pH, including KatG. In addition, heme peroxidases such as KatG oxidize nitrite to more reactive species, such as nitrogen dioxide (•NO2) and nitryl chloride (NO2Cl) (Klebanoff 1993; van der Vliet et al. 1997; Eiserich et al. 1998; Battistuzzi et al. 2010).
INH is widely reported to lose mycobactericidal activity when Mtb is cultured in conditions that prevent it from replicating. However, when the oxygen in an axenic, nitrate-containing culture of Mtb was reduced below 1%, narG-deficient Mtb, which was not replicating, became highly INH sensitive. We confirmed that effect and demonstrated its reversal with exogenous nitrite. To the extent that a relevant source of nitrite for Mtb residing in a macrophage in the human host may be Mtb itself, the question arises whether the cidality of INH for nonreplicating narG-deficient Mtb could be phenocopied in nonreplicating wild-type Mtb by an inhibitor of Mtb's nitrate transporter or nitrate reductase. If so, such agents might significantly reduce the time required to cure latent TB with INH or active TB with INH-containing regimens, reducing the incidence of emergent drug resistance associated with incomplete treatment (van den Boogaard et al. 2009; Zumla et al. 2013). InhA is essential to aerobically and perhaps also hypoxically cultured mycobacteria as two compounds that targeted both InhA as well as fatty acid synthase type 1 were cidal to nonreplicating Mtb (Vilcheze et al. 2011a,b). Nevertheless, it remains unclear whether the reduced survival of narG-deficient Mtb treated with INH resulted only from the inactivation of InhA or by inhibition of additional mycobacterial target(s). A mechanistic understanding of the hypersusceptibility of narG-deficient Mtb to INH may reveal new INH target(s) amenable to inhibition by other compounds.
Acknowledgments
We are grateful to the blood donors for their participation. We thank Guillaume Vogt for instruction in the human macrophage culture system and his original observation of nitrite accumulation in the coculture. We appreciate the technical assistance provided by Xiuju Jiang and the anti-DLAT antibody from Ruslana Bryk. We thank Kristin Burns for guidance and Stefan Ehlers, Ruslana Bryk, Ben Gold, Sabine Ehrt, Dirk Schnappinger, Poonam Rath, Michele Fuortes, and K. Heran Darwin for advice. ACB was supported by a Medical Scientist Training Program grant from the National Institute of General Medical Sciences of the National Institutes of Health under award number GM07739 to the Weill Cornell/Rockefeller/Sloan-Kettering Tri-Institutional MD-PhD Program. The Department of Microbiology and Immunology is supported by the William Randolph Hearst Foundation.
Conflict of Interest
None declared.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Figure S1. (A–C) Survival of wild-type, narG-deficient (NarG), and complemented strains (NarGc) within macrophages (MOI: 0.1, corresponding to 10,000 bacteria) treated with the indicated concentrations of (A) rifampicin (B) streptomycin, or (C) ethambutol over 3 days in 10% oxygen. The results shown in A–C are single experiments that are representative of at least two independent experiments conducted using cells isolated from at least two distinct human donors. (D–F) contain the pooled CFU data from at least two independent experiments conducted with cells from at least two individual human donors shown for one drug concentration as follows: (D) rifampicin (0.2 μg/mL), (E) streptomycin (0.6 μg/mL), or (F) ethambutol (0.1 μg/mL). Bars indicate means ± SEM.
Figure S2. (A) The survival of axenic cultures of Mtb (OD 0.1) following treatment with the indicated concentration of hydrogen peroxide at 1% oxygen incubated overnight with 1 mmol/L nitrite. This is representative of two independent experiments. (B) The mRNA expression of katG by wild-type, narG -deficient (NarG), and the complemented (NarGc) strains within infected human macrophages following incubation for 3 days at 10% oxygen. The given experiment is the pooled result of two independent experiments. (C) Axenic cultures of wild-type, narG-deficient, and the complemented strains of Mtb were incubated over 1 day at 1% oxygen and lysed to collect total cell protein. Individual proteins from the total cell lysate were separated by native gel electrophoresis and KatG and the housekeeping gene DLAT were visualized by fluorophore-coupled antibody staining. This is representative of three independent experiments. Bars indicate means ± SEM.
References
- Angeby KAK, Klintz L, Hoffner SE. Rapid and inexpensive drug susceptibility testing of Mycobacterium tuberculosis with a nitrate reductase assay. J. Clin. Microbiol. 2002;40:553–555. doi: 10.1128/JCM.40.2.553-555.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battistuzzi G, Bellei M, Bortolotti CA, Sola M. Redox properties of heme peroxidases. Arch. Biochem. Biophys. 2010;500:21–36. doi: 10.1016/j.abb.2010.03.002. [DOI] [PubMed] [Google Scholar]
- Baulard AR, Betts JC, Engohang-Ndong J, Quan S, McAdam RA, Brennan PJ, et al. Activation of the pro-drug ethionamide is regulated in mycobacteria. J. Biol. Chem. 2000;275:28326–28331. doi: 10.1074/jbc.M003744200. [DOI] [PubMed] [Google Scholar]
- van den Boogaard J, Lyimo R, Irongo CF, Boeree MJ, Schaalma H, Aarnoutse RE, et al. Community vs. facility-based directly observed treatment for tuberculosis in Tanzania's Kilimanjaro Region. Int. J. Tuberc. Lung Dis. 2009;13:1524–1529. [PubMed] [Google Scholar]
- Cade CE, Dlouhy AC, Medzihradszky KF, Salas-Castillo SP, Ghiladi RA. Isoniazid-resistance conferring mutations in Mycobacterium tuberculosis KatG: catalase, peroxidase, and INH-NADH adduct formation activities. Protein Sci. 2010:458–474. doi: 10.1002/pro.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H-S, Rai PR, Chu HW, Cool C, Chan ED. Analysis of nitric oxide synthase and nitrotyrosine expression in human pulmonary tuberculosis. Am. J. Respir. Crit. Care Med. 2002;166:178–186. doi: 10.1164/rccm.2201023. [DOI] [PubMed] [Google Scholar]
- Corker H. Nitric oxide formation by Escherichia coli: dependence on nitrite reductase, the no-sensing regulator Fnr, and flavohemoglobin Hmp. J. Biol. Chem. 2003;278:31584–31592. doi: 10.1074/jbc.M303282200. [DOI] [PubMed] [Google Scholar]
- DeBarber AE, Mdluli K, Bosman M, Bekker LG, Barry CE. Ethionamide activation and sensitivity in multidrug-resistant Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA. 2000;97:9677–9682. doi: 10.1073/pnas.97.17.9677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar N, McKinney JD. Mycobacterium tuberculosis persistence mutants identified by screening in isoniazid-treated mice. Proc. Natl. Acad. Sci. USA. 2010;107:12275–12280. doi: 10.1073/pnas.1003219107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiserich JP, Hristova M, Cross CE, Jones AD, Freeman BA, Halliwell B, et al. Formation of nitric oxide-derived inflammatory oxidants by myeloperoxidase in neutrophils. Nature. 1998;391:393–397. doi: 10.1038/34923. [DOI] [PubMed] [Google Scholar]
- Fenhalls G, Stevens L, Moses L, Bezuidenhout J, Betts JC, Helden Pv PV, et al. In situ detection of Mycobacterium tuberculosis transcripts in human lung granulomas reveals differential gene expression in necrotic lesions. Infect. Immun. 2002;70:6330–6338. doi: 10.1128/IAI.70.11.6330-6338.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiladi RA, Medzihradszky KF, Rusnak FM, Ortiz de Montellano PR. Correlation between isoniazid resistance and superoxide reactivity in Mycobacterium tuberculosis KatG. J. Am. Chem. Soc. 2005;127:13428–13442. doi: 10.1021/ja054366t. [DOI] [PubMed] [Google Scholar]
- Giffin MM, Raab RW, Morganstern M, Sohaskey CD. Mutational analysis of the respiratory nitrate transporter NarK2 of Mycobacterium tuberculosis. PLoS One. 2012;7:e45459. doi: 10.1371/journal.pone.0045459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazbon MH, Brimacombe M, Bobadilla del Valle M, Cavatore M, Guerrero MI, Varma-Basil M, et al. Population genetics study of isoniazid resistance mutations and evolution of multidrug-resistant Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2006;50:2640–2649. doi: 10.1128/AAC.00112-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebanoff SJ. Reactive nitrogen intermediates and antimicrobial activity: role of nitrite. Free Radic. Biol. Med. 1993;14:351–360. doi: 10.1016/0891-5849(93)90084-8. [DOI] [PubMed] [Google Scholar]
- Lundberg JO, Weitzberg E. NO-synthase independent NO generation in mammals. Biochem. Biophys. Res. Commun. 2010;396:39–45. doi: 10.1016/j.bbrc.2010.02.136. [DOI] [PubMed] [Google Scholar]
- Lundberg JO, Weitzberg E, Gladwin MT. The nitrate–nitrite–nitric oxide pathway in physiology and therapeutics. Nat. Rev. Drug Discov. 2008;7:156–167. doi: 10.1038/nrd2466. [DOI] [PubMed] [Google Scholar]
- Lundberg JO, Gladwin MT, Ahluwalia A, Benjamin N, Bryan NS, Butler A, et al. Nitrate and nitrite in biology, nutrition and therapeutics. Nat. Chem. Biol. 2009;5:865–869. doi: 10.1038/nchembio.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musser JM, Kapur V, Williams DL, Kreiswirth BN, van Soolingen D, van Embden JD. Characterization of the catalase-peroxidase gene (katG) and inhA locus in isoniazid-resistant and -susceptible strains of Mycobacterium tuberculosis by automated DNA sequencing: restricted array of mutations associated with drug resistance. J. Infect. Dis. 1996;173:196–202. doi: 10.1093/infdis/173.1.196. [DOI] [PubMed] [Google Scholar]
- Nathan C, Shiloh MU. Reactive oxygen and nitrogen intermediates in the relationship between mammalian hosts and microbial pathogens. Proc. Natl. Acad. Sci. USA. 2000;97:8841–8848. doi: 10.1073/pnas.97.16.8841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson S, Bonecini-Almeida MDG, Lapa e Silva JR, Nathan C, Xie QW, Mumford R, et al. Inducible nitric oxide synthase in pulmonary alveolar macrophages from patients with tuberculosis. J. Exp. Med. 1996;183:2293–2302. doi: 10.1084/jem.183.5.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacelli R, Wink DA, Cook JA, Krishna MC, DeGraff W, Friedman N, et al. Nitric oxide potentiates hydrogen peroxide-induced killing of Escherichia coli. J. Exp. Med. 1995;182:1469–1479. doi: 10.1084/jem.182.5.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachman H, Strong M, Ulrichs T, Grode L, Schuchhardt J, Mollenkopf H, et al. Unique transcriptome signature of Mycobacterium tuberculosis in pulmonary tuberculosis. Infect. Immun. 2006;74:1233–1242. doi: 10.1128/IAI.74.2.1233-1242.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralt D, Wishnok JS, Fitts R, Tannenbaum SR. Bacterial catalysis of nitrosation: involvement of the nar operon of Escherichia coli. J. Bacteriol. 1988;170:359–364. doi: 10.1128/jb.170.1.359-364.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee KY, Erdjument-Bromage H, Tempst P, Nathan CF. S-nitroso proteome of Mycobacterium tuberculosis: enzymes of intermediary metabolism and antioxidant defense. Proc. Natl. Acad. Sci. USA. 2005;102:467–472. doi: 10.1073/pnas.0406133102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozwarski DA. Modification of the NADH of the isoniazid target (InhA) from Mycobacterium tuberculosis. Science. 1998;279:98–102. doi: 10.1126/science.279.5347.98. [DOI] [PubMed] [Google Scholar]
- Saint-Joanis B, Souchon H, Wilming M, Johnsson K, Alzari PM, Cole ST. Use of site-directed mutagenesis to probe the structure, function and isoniazid activation of the catalase/peroxidase, KatG, from Mycobacterium tuberculosis. Biochem. J. 1999;338:753–760. [PMC free article] [PubMed] [Google Scholar]
- Seth D, Hausladen A, Wang YJ, Stamler JS. Endogenous protein S-Nitrosylation in E. coli: regulation by OxyR. Science. 2012;336:470–473. doi: 10.1126/science.1215643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman DR, Voskuil M, Schnappinger D, Liao R, Harrell MI, Schoolnik GK. Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding alpha -crystallin. Proc. Natl. Acad. Sci. USA. 2001;98:7534–7539. doi: 10.1073/pnas.121172498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Sohaskey CD, Kana BD, Dawes S, North RJ, Mizrahi V, et al. Changes in energy metabolism of Mycobacterium tuberculosis in mouse lung and under in vitro conditions affecting aerobic respiration. Proc. Natl Acad. Sci. USA. 2005;102:15629–15634. doi: 10.1073/pnas.0507850102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohaskey CD, Wayne LG. Role of narK2X and narGHJI in hypoxic upregulation of nitrate reduction by Mycobacterium tuberculosis. J. Bacteriol. 2003;185:7247–7256. doi: 10.1128/JB.185.24.7247-7256.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stermann M, Bohrssen A, Diephaus C, Maass S, Bange F-C. Polymorphic nucleotide within the promoter of nitrate reductase (NarGHJI) is specific for Mycobacterium tuberculosis. J. Clin. Microbiol. 2003;41:3252–3259. doi: 10.1128/JCM.41.7.3252-3259.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venugopal A, Bryk R, Shi S, Rhee K, Rath P, Schnappinger D, et al. Virulence of Mycobacterium tuberculosis depends on lipoamide dehydrogenase, a member of three multienzyme complexes. Cell Host Microbe. 2011;9:21–31. doi: 10.1016/j.chom.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilcheze C, Av-Gay Y, Barnes SW, Larsen MH, Walker JR, Glynne RJ, et al. Coresistance to isoniazid and ethionamide maps to mycothiol biosynthetic genes in Mycobacterium bovis. Antimicrob. Agents Chemother. 2011a;55:4422–4423. doi: 10.1128/AAC.00564-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilcheze C, Baughn AD, Tufariello J, Leung LW, Kuo M, Basler CF, et al. Novel inhibitors of InhA efficiently kill Mycobacterium tuberculosis under aerobic and anaerobic conditions. Antimicrob. Agents Chemother. 2011b;55:3889–3898. doi: 10.1128/AAC.00266-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilchèze C, Weisbrod TR, Chen B, Kremer L, Hazbón MH, Wang F, et al. Altered NADH/NAD+ ratio mediates coresistance to isoniazid and ethionamide in mycobacteria. Antimicrob. Agents Chemother. 2005;49:708–720. doi: 10.1128/AAC.49.2.708-720.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vliet A, Eiserich JP, Halliwell B, Cross CE. Formation of reactive nitrogen species during peroxidase-catalyzed oxidation of nitrite. A potential additional mechanism of nitric oxide-dependent toxicity. J. Biol. Chem. 1997;272:7617–7625. doi: 10.1074/jbc.272.12.7617. [DOI] [PubMed] [Google Scholar]
- Vogt G, Nathan C. In vitro differentiation of human macrophages with enhanced antimycobacterial activity. J. Clin. Invest. 2011;121:3889–3901. doi: 10.1172/JCI57235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voskuil MI, Schnappinger D, Visconti KC, Harrell MI, Dolganov GM, Sherman DR, et al. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J. Exp. Med. 2003;198:705–713. doi: 10.1084/jem.20030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanger A, Mills K. Testing of Mycobacterium tuberculosis susceptibility to ethambutol, isoniazid, rifampin, and streptomycin by using Etest. J. Clin. Microbiol. 1996;34:1672–1676. doi: 10.1128/jcm.34.7.1672-1676.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne LG, Hayes LG. Nitrate reduction as a marker for hypoxic shiftdown of Mycobacterium tuberculosis. Tuber. Lung Dis. 1998;79:127–132. doi: 10.1054/tuld.1998.0015. [DOI] [PubMed] [Google Scholar]
- Wayne LG, Sohaskey CD. Nonreplicating persistence of Mycobacterium tuberculosis. Annu. Rev. Microbiol. 2001;55:139–163. doi: 10.1146/annurev.micro.55.1.139. [DOI] [PubMed] [Google Scholar]
- Wengenack NL, Lane BD, Hill PJ, Uhl JR, Lukat-Rodgers GS, Hall L, et al. Purification and characterization of Mycobacterium tuberculosis KatG, KatG(S315T), and Mycobacterium bovis KatG(R463L) Protein Expr. Purif. 2004;36:232–243. doi: 10.1016/j.pep.2004.04.018. [DOI] [PubMed] [Google Scholar]
- Wilson M, DeRisi J, Kristensen HH, Imboden P, Rane S, Brown PO, et al. Exploring drug-induced alterations in gene expression in Mycobacterium tuberculosis by microarray hybridization. Proc. Natl Acad. Sci. USA. 1999;96:12833–12838. doi: 10.1073/pnas.96.22.12833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winder FG, Collins PB. Inhibition by isoniazid of synthesis of mycolic acids in Mycobacterium tuberculosis. J. Gen. Microbiol. 1970;63:41–48. doi: 10.1099/00221287-63-1-41. [DOI] [PubMed] [Google Scholar]
- Woodmansee AN, Imlay JA. A mechanism by which nitric oxide accelerates the rate of oxidative DNA damage in Escherichia coli. Mol. Microbiol. 2003;49:11–22. doi: 10.1046/j.1365-2958.2003.03530.x. [DOI] [PubMed] [Google Scholar]
- Zumla A, Raviglione M, Hafner R, Fordham von Reyn C. Tuberculosis. N. Engl. J. Med. 2013;368:745–755. doi: 10.1056/NEJMra1200894. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.