Figure 2. Peak Statistics, Motif Analyses and GREAT annotations of β-catenin and Six2 ChIP-seq datasets.
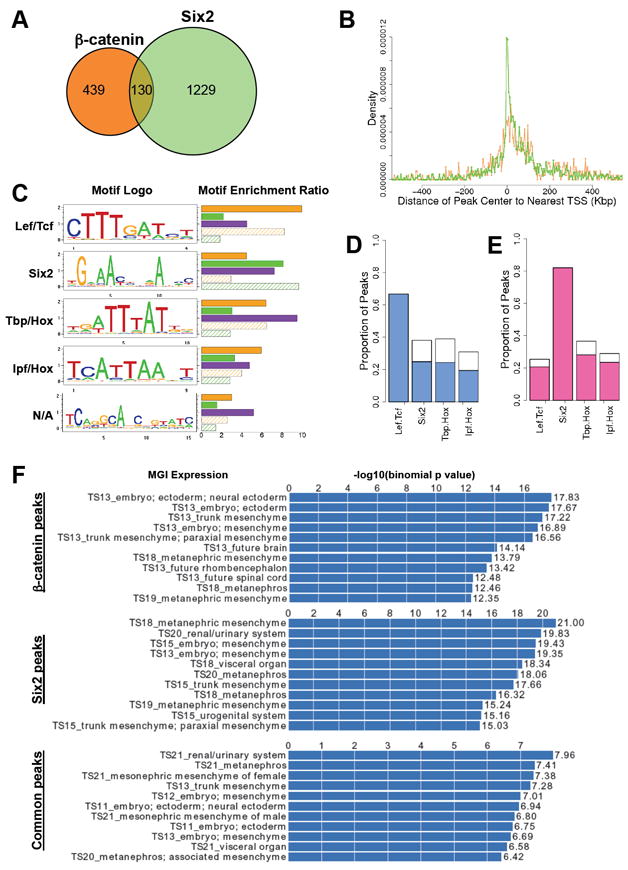
(A) Venn-diagram of the overlap between β-catenin (orange circle) and Six2 (green circle) bound ChIP products with a 0.01 FDR cutoff.
(B) Density graph of the peak locations in relation to the nearest transcriptional start sites (TSS). X-axis represents distance from the peak center to the nearest TSS, where 0 is the position of the TSS. Y-axis represents proportion of detected bindings that are located within each distance interval (distance increasing at 10 kb intervals). Orange line, β-catenin peaks; Green line, Six2 peaks.
(C) Top five enriched de novo motifs recovered from ChIP-seq peak regions. Separate runs of de novo motif discovery were performed on genomic regions bound by β-catenin, Six2, or both. The motif logos display nucleotide frequencies (scaled relative to the information content) at each position. The horizontal bars on the right side represent the motif enrichment in peak regions (r3, defined in Supplemental Experimental Procedures). For each motif, we calculate its enrichment in five different sets of peak regions: β-catenin peak regions (orange bar), Six2 peak regions (green bar), common peak regions (purple bar), β-catenin distinctive peak regions (orange shadowed bar) and Six2 distinctive peak regions (green shadowed bar). N/A, not applicable.
(D) De novo motif occupancies in β-catenin peak regions. The blue bars are for the peaks containing de novo Lef/Tcf motif sites.
(E) De novo motif occupancies in Six2 peak regions. The pink bars are for the peaks containing de novo Six2 motif sites.
(F) MGI expression annotations of β-catenin, Six2 and common peaks analyzed by GREAT. A sample of 11 representative annotations are shown for each peak category. Histogram represents the –log10 (binomial p-value) for each annotation.
