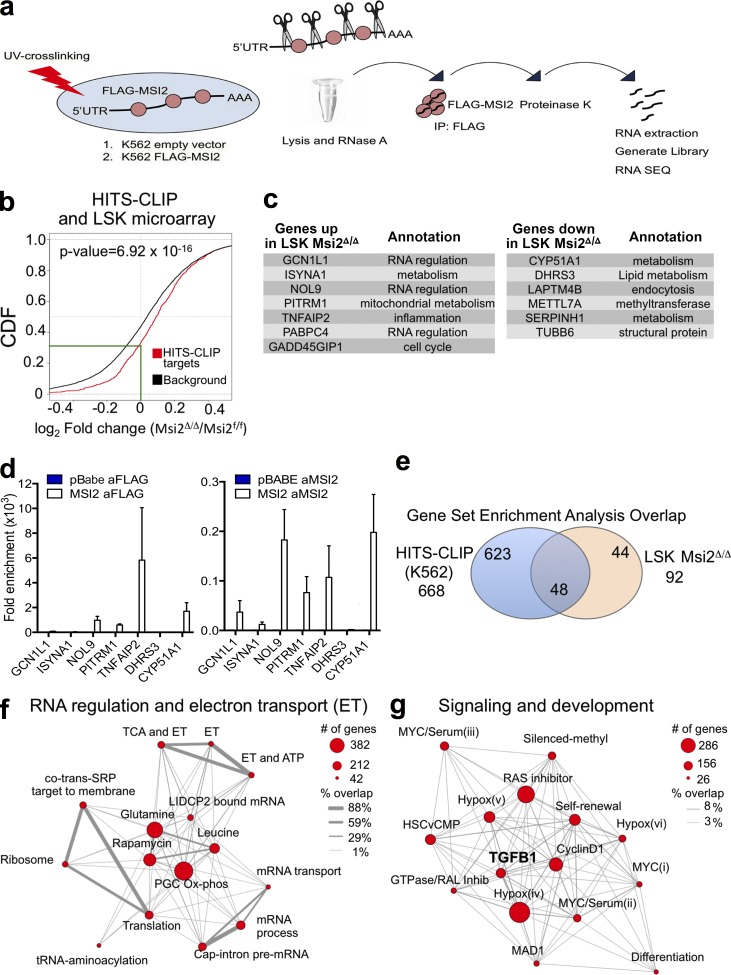
Figure 7.
MSI2’s direct RNA targets are involved in RNA metabolism and the self-renewal signaling program. (a) Schematic outline of the HITS-CLIP approach. (b) Target genes are mouse genes homologous to human MSI2 targets identified by HITS-CLIP in K562 (FDR < 0.1) and background genes are all the other genes expressed in mouse LSK cells. Log2 fold changes (logFCs) of gene expression between knockout and wild-type samples are fitted using limma package. When there are multiple microarray probes mapped to the same gene, logFC of that gene is represented by the median of all corresponding probes. P-value is obtained with one-sided K-S test on the logFC distributions of target and background genes (green line indicates the intersection of the HITS-CLIP targets with a zero logFC). CDF, cumulative distribution function. (c) Up- and down-regulated genes that are differentially expressed and overlap with the top HITS-CLIP targets. (d) Validation using qRT-PCR of a subset of the HITS-CLIP targets from c with RIP performed with anti-FLAG Ab and Ab specific for MSI2 in K562 cells (mean of two independent RIP experiments and fold change over control samples; pBabe cells were used as control for Flag Ab sample, and anti–rabbit Ab was used as control for Msi2 Ab). SEM is shown. (e) Venn diagram indicating the number of statistically significant gene sets from the HITS-CLIP or the transcriptome analysis comparing controls and Msi2Δ/Δ LSKs (FDR < 0.01). The number in the middle indicates the overlapping significant gene sets that are enriched in both the HITS-CLIP and the transcriptome analysis of LSKs. (f and g) Modules indicate the relationships between manually curated and selected gene sets (within the overlap in e and Table S6). “% overlap” indicates the number of genes that are common between the genes set.