The transcription factor Stat5 inhibits the bone-resorbing function of osteoclasts, in part by inducing the expression of the phosphatases Dusp1 and Dusp2.
Abstract
Signal transducer and activator of transcription 5 (Stat5) is essential for cytokine-regulated processes such as proliferation, differentiation, and survival in hematopoietic cells. To investigate the role of Stat5 in osteoclasts, we generated mice with an osteoclast-specific conditional deletion of Stat5 (Stat5 conditional knockout [cKO] mice) and analyzed their bone phenotype. Stat5 cKO mice exhibited osteoporosis caused by an increased bone-resorbing activity of osteoclasts. The activity of mitogen-activated protein kinases (MAPKs), in particular extracellular signal–related kinase, was increased in Stat5 cKO osteoclasts, whereas the expression of the MAPK phosphatases dual specificity phosphatase 1 (Dusp1) and Dusp2 was significantly decreased. Interleukin-3 (IL-3) stimulated the phosphorylation and nuclear translocation of Stat5 in osteoclasts, and Stat5 expression was up-regulated in response to receptor activator of nuclear factor κB ligand (RANKL). The results suggest that Stat5 negatively regulates the bone-resorbing function of osteoclasts by promoting Dusp1 and Dusp2 expression, and IL-3 promotes Stat5 activation in osteoclasts.
Bone is a dynamic tissue that undergoes constant remodeling, balancing bone resorption by osteoclasts with bone formation by osteoblasts (Baron, 1989; Karsenty and Wagner, 2002). An altered balance between bone formation and bone resorption causes diseases such as osteoporosis and osteopetrosis (Seeman, 2003; Villa et al., 2009). It is well known that osteoclasts are responsible for the pathological bone resorption in various diseases, including rheumatoid arthritis, the loosening of artificial joints, and osteolysis in metastatic cancers (Takayanagi et al., 2000; Roodman, 2004; Sommer et al., 2005; Tanaka, 2007). Osteoclasts are multinucleated bone-resorbing cells derived from hematopoietic precursors of the monocyte-macrophage lineage, and their differentiation is induced by M-CSF and receptor activator of NF-κB ligand (RANKL; Lacey et al., 1998; Yasuda et al., 1998; Suda et al., 1999). RANKL is not only important for the differentiation of osteoclasts, but it also regulates their bone-resorbing activity and survival. Although bone resorption is tightly regulated in the skeletal milieu, the molecular mechanisms underlying it are not fully understood.
The Stat family of transcription factors conveys cytokine signals from the respective membrane receptors to the nucleus, where they activate diverse genetic programs (Akira, 1999). Stat5 was originally identified as mammary gland factor (MGF), implying a specificity to the physiology of mammary tissue (Wakao et al., 1994). Two members of the Stat family, Stat5a and Stat5b (collectively called Stat5), have gained prominence in that they are activated by a wide variety of cytokines (Hennighausen and Robinson, 2008). Stat5a and Stat5b play redundant and nonredundant roles that are essential in a variety of cytokine responses (Teglund et al., 1998). Recently, it was demonstrated that Stat5 has important functions in immune cell development. Mice completely lacking Stat5 failed to develop T or B lymphocytes as well as NK cells (Hoelbl et al., 2006; Yao et al., 2006), and Stat5-null embryos are perinatally lethal, possibly as the result of severe anemia (Cui et al., 2004). Stat5 also plays an essential role in certain pathological conditions. Activated STAT5 was detected in BCR-ABL–induced chronic myeloid leukemia (Ilaria and Van Etten, 1996), and a recent study reported that activated Stat5 was detected in the blast cells of acute myeloblastic leukemia and acute lymphoblastic leukemia patients (Van Etten, 2007). These results suggest that Stat5 plays an essential role in a variety of cells, particularly of the hematopoietic lineage. However, the role of Stat5 in osteoclasts has yet to be elucidated.
In this study, we investigated the roles of Stat5 in osteoclasts in vivo and in vitro. It was demonstrated that Stat5 negatively regulates the bone-resorbing activity of osteoclasts and that osteoclast-specific Stat5 KO mice exhibited reduced bone mass. We identified dual specificity mitogen-activated protein kinase (MAPK) phosphatase 1 (Dusp1) and Dusp2 as targets of Stat5. Only IL-3 among the known stimulators activated Stat5 and induced the expression of both Dusp1 and Dusp2. IL-3 expression is up-regulated in osteoclasts in response to RANKL, thus providing a cell-autonomous negative feedback loop in bone resorption.
RESULTS
Stat5 conditional KO (cKO) mice exhibit decreased bone mass
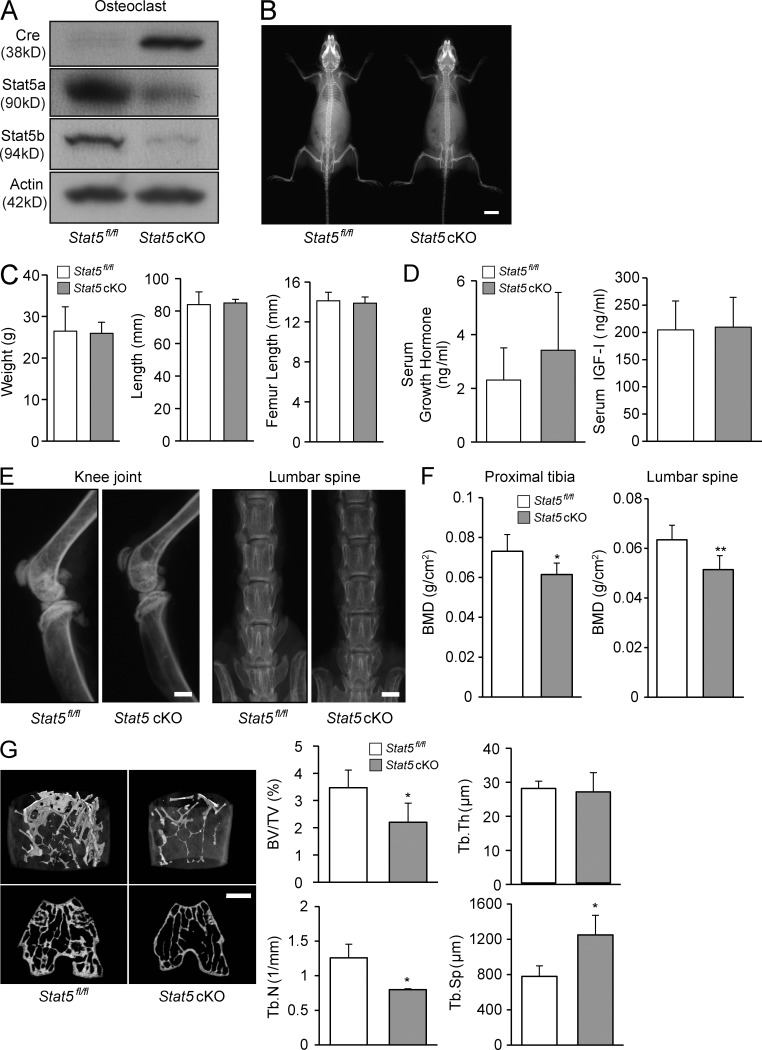
To investigate the role of Stat5 in osteoclasts, we generated osteoclast-specific Stat5a&b cKO (Stat5 cKO) mice by mating Stat5fl/fl mice (Cui et al., 2004) with cathepsin K–Cre transgenic mice, in which the Cre recombinase gene is inserted into the cathepsin K locus and specifically expressed in osteoclasts (Nakamura et al., 2007). Stat5 cKO mice were born alive at predicted Mendelian frequencies. Stat5 expression was barely observed in osteoclasts from the Stat5 cKO mice (Fig. 1 A), whereas its expression in other tissues was comparable with that in the Stat5fl/fl littermates (not depicted). Stat5 cKO mice grew normally with no apparent morphological abnormalities (Fig. 1 B) and they exhibited a normal height and weight (Fig. 1 C). In addition, the serum levels of growth hormone or insulin like growth factor I (IGF-I) in Stat5 cKO mice were within normal range and exhibited no significant difference from normal littermates (Fig. 1 D). 1-yr-old male Stat5 cKO mice exhibited a decreased bone mass in the proximal tibia and lumbar spine, as shown by radiography (Fig. 1 E) and dual-energy x-ray absorptiometry (DXA; Fig. 1 F). Microcomputed tomography (micro-CT) analysis of the distal femur revealed apparent osteopenia in the Stat5 cKO male mice at 1 yr of age. Bone volume/tissue volume (BV/TV) and trabecular number (Tb.N) were significantly reduced, and trabecular separation (Tb.Sp) significantly increased in 1-yr-old Stat5 cKO mice compared with Stat5fl/fl mice (Fig. 1 G). These data suggest that Stat5 signaling plays an important role in regulating bone homeostasis.
Figure 1.
Stat5 cKO mice exhibit decreased bone mass. (A) Expression of Cre recombinase and Stat5 in osteoclasts from Stat5fl/fl mice and Stat5 cKO mice was analyzed by Western blotting. β-Actin was used as an internal control. (B) Radiographs of a Stat5fl/fl male mouse (left) and a Stat5 cKO male mouse (right) at 3 mo of age. (C) Body weight and length and femur length were measured for 12-wk-old Stat5fl/fl and Stat5 cKO male mice. Data are the mean ± SD (n = 4). (D) Serum levels of growth hormone (left) and IGF-I (right) in Stat5fl/fl mice and Stat5 cKO mice were measured by ELISA. Data are represented as the mean ± SD (n = 6). (E) Representative x-ray images of the knee joint and lumbar spine of a Stat5fl/fl male mouse (left) and a Stat5 cKO male mouse (right) at 1 yr of age. (F) Bone mineral density (BMD) of the distal femur and lumbar spine of 1-yr-old Stat5fl/fl male mice and Stat5 cKO male mice was determined by DXA. Data are the mean ± SD (*, P < 0.05; **, P < 0.01; n = 6). (G) Representative micro-CT images of the distal femur in a Stat5fl/fl mouse (left) and Stat5 cKO mouse (right) at 1 yr of age. Graphs show quantitative data from the CT analyses. BV/TV, bone volume per total volume; Tb.N, trabecular number; Tb.Th, trabecular thickness; Tb.Sp, trabecular separation. Data are represented as the mean ± SD (*, P < 0.05; n = 3). Bars: (B) 1 cm; (E and G) 1 mm.
Bone resorption is increased in Stat5 cKO mice
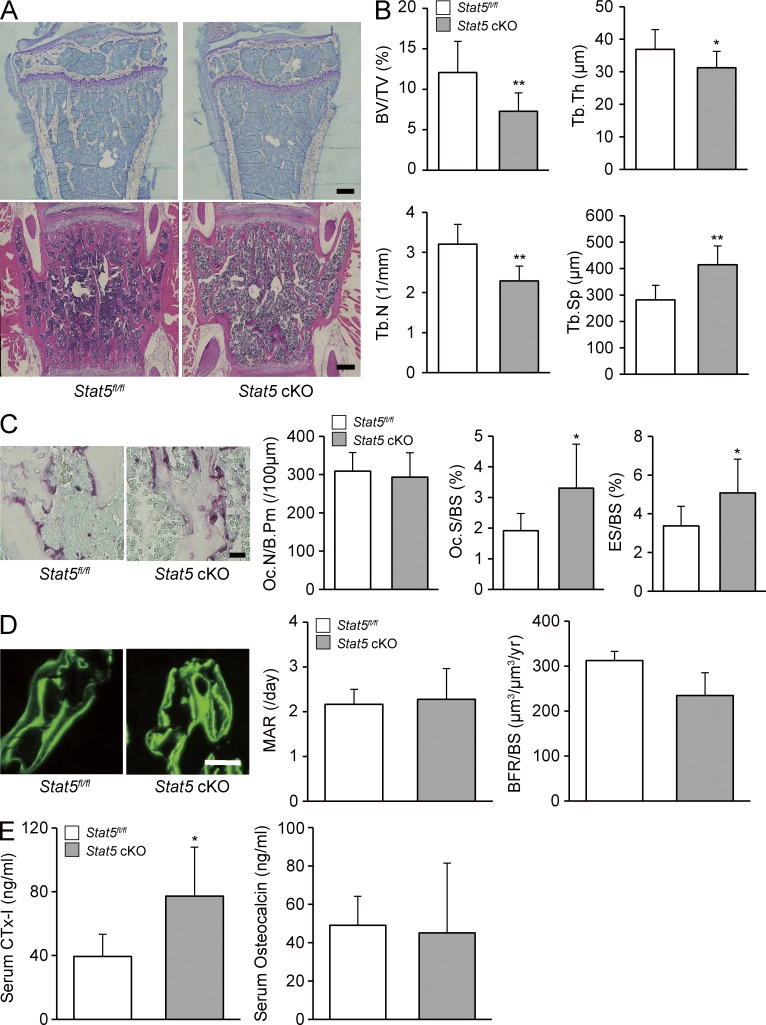
Histological and histomorphometric analyses revealed that a decrease in trabecular bone volume was already taking place in 8-wk-old Stat5 cKO male mice compared with Stat5fl/fl mice (Fig. 2, A and B). 8-wk-old Stat5 cKO female mice exhibited a significantly decreased Tb.N and an increased Tb.Sp, whereas BV/TV was not significantly different between cKO mice and control mice (not depicted). The Stat5 cKO male mice exhibited a significant increase in the eroded surface/bone surface (ES/BS) and osteoclast surface/bone surface (Oc.S/BS), but not in osteoclast number (Oc.N/B.Pm), as shown by tartrate-resistant acid phosphatase (TRAP) staining of bone sections and histomorphometric analysis (Fig. 2 C). In contrast, the bone formation parameters, mineral apposition rate (MAR) and bone formation rate (BFR/BS), did not exhibit any significant difference between Stat5 cKO mice and Stat5fl/fl mice (Fig. 2 D). The serum levels of C-terminal cross-linking telopeptide of type 1 collagen (CTx-I), a bone resorption marker, were significantly increased in Stat5 cKO mice compared with Stat5fl/fl mice, whereas the serum levels of osteocalcin, a bone formation marker, were equivalent to those in Stat5fl/fl mice (Fig. 2 E). These findings suggest that bone loss in Stat5 cKO mice is caused by an increased bone-resorbing activity of osteoclasts, rather than by decreased bone formation.
Figure 2.
Bone resorption was promoted in Stat5 cKO mice. (A) Undecalcified sections of the proximal tibia (top) or decalcified section of the lumbar spine (bottom) from 8-wk-old Stat5fl/fl male mice (left) or Stat5 cKO male mice (right) were stained with toluidine blue (top) or H&E (bottom). Representative images from three mice/group are shown. (B) Histomorphometric analyses were performed on 8-wk-old Stat5fl/fl male mice and Stat5 cKO male mice. BV/TV, bone volume per total volume; Tb.Th, trabecular thickness; Tb.N, trabecular number; Tb.Sp, trabecular separation. Data are represented as the mean ± SD (*, P < 0.05; **, P < 0.01; n = 6). (C) TRAP expression in tibia from Stat5fl/fl (left) or Stat5 cKO mice (right) was assessed by TRAP staining and the following parameters were measured: Oc.N/B.Pm, osteoclast number per bone perimeter; Oc.S/BS, osteoclast surface per bone surface; ES/BS, eroded surface per bone surface. Data are represented as the mean ± SD (*, P < 0.05; n = 6). (D) Dynamic histomorphometric analysis of Stat5fl/fl and Stat5 cKO mice was performed after mice were injected subcutaneously with 16 mg/kg body weight of calcein at −3 and −1 d before sacrifice. Graphs show MAR, mineral apposition rate; BFR/BS, bone formation rate per bone surface. Data are represented as the mean ± SD (n = 6). (E) Serum concentration of CTx-I and osteocalcin in Stat5fl/fl mice and Stat5 cKO mice was assessed by ELISA. Data are represented as the mean ± SD (*, P < 0.05; n = 4/group). Bars: (A and D) 100 µm; (C) 25 µm.
Stat5 negatively regulates osteoclast function
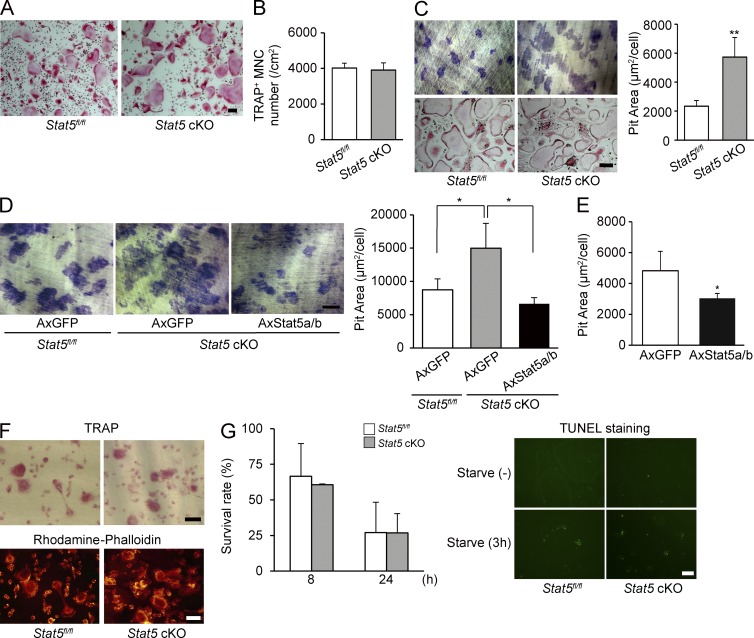
To clarify the mechanism of increased bone resorption by Stat5 deletion, we performed in vitro experiments using BM cells isolated from Stat5 cKO and Stat5fl/fl mice. We first evaluated whether the M-CSF–dependent proliferation of BM cells was different between Stat5 cKO and Stat5fl/fl mice by MTT assay. Proliferation was comparable between cells from Stat5fl/fl mice and those from Stat5 cKO mice (not depicted). The RANKL-induced osteoclastic differentiation of BM cells from Stat5 cKO mice did not differ from cells from Stat5fl/fl mice, as demonstrated by TRAP staining (Fig. 3 A), the TRAP+ multinucleated cell number (Fig. 3 B), and the expression of osteoclast differentiation markers (Nfatc1, Ctsk, and Acp5; not depicted). We next evaluated the bone-resorbing activity of fully differentiated osteoclasts by pit formation assay on dentine slices. Osteoclasts differentiated from the BM cells of the Stat5 cKO mice (Stat5 cKO osteoclasts) exhibited an increased bone-resorbing activity compared with osteoclasts from the Stat5fl/fl mice (Stat5fl/fl osteoclasts; Fig. 3 C). The increased bone-resorbing activity of Stat5 cKO osteoclasts was hindered by the adenoviral reintroduction of Stat5a/b (Fig. 3 D). Inversely, Stat5 overexpression significantly reduced the bone-resorbing activity of osteoclasts (Fig. 3 E). The size or actin organization did not differ apparently between Stat5 cKO and Stat5fl/fl osteoclasts on dentin slices, though the size is quite different between individual osteoclast (Fig. 3 F). We then generated osteoclasts from Stat5 cKO mice BM cells by stimulating these cells with M-CSF and RANKL and examined the effect of Stat5 deficiency on osteoclast survival. As shown in Fig. 3 G, the loss of Stat5 did not affect the osteoclast survival rate or apoptosis of osteoclasts evaluated by TUNEL staining. The expression of cleaved caspase-3, a primary caspase in the execution of apoptosis, was also not significantly different (not depicted). These results suggest that Stat5 is essential for the bone-resorbing activity but not the differentiation or survival of osteoclasts.
Figure 3.
Stat5 suppresses the bone-resorbing activity of osteoclasts. (A) BMDMs from Stat5fl/fl mice and Stat5 cKO mice were cultured in the presence of M-CSF and RANKL for 4 d, and cultures were stained for TRAP+ cells. (B) BMDMs from Stat5fl/fl mice and Stat5 cKO mice were cultured in the presence of M-CSF and RANKL for 4 d, and the TRAP+ multinucleated cells (MNCs) containing more than three nuclei were counted as osteoclasts. Data are represented as the mean ± SD (n = 3). (C) Osteoclasts from Stat5fl/fl and Stat5 cKO mice were cultured on dentine slices for 24 h, and the resorption areas were visualized by staining with toluidine blue staining and measured using an image analysis system. Representative resorption pits and TRAP staining of parallel cultures are shown (left). Graph shows pit resorption area per osteoclast (right). Data are represented as the mean ± SD (**, P < 0.01; n = 3). (D) Osteoclasts from Stat5fl/fl or Stat5 cKO mice infected with AxGFP or AxStat5a/b were cultured on dentine slices for 24 h, and the resorption areas were visualized by staining with toluidine blue staining. Pit resorption area per osteoclast and representative resorption pits are shown. Data are represented as the mean ± SD (*, P < 0.05; n = 3). (E) Osteoclasts from WT mice infected with AxGFP or AxStat5a/b were cultured on dentine slices for 24 h, and the resorption areas were visualized by staining with toluidine blue staining. Graph shows pit resorption area per osteoclast. Data are represented as the mean ± SD (*, P < 0.05; n = 3). (F) TRAP staining and rhodamine-conjugated phalloidin staining of osteoclasts from Stat5fl/fl mice and Stat5 cKO mice on dentin slices. (G) Stat5fl/fl and Stat5 cKO osteoclasts were cultured for 8 or 24 h after cytokine withdrawal (left). Apoptosis of osteoclasts from Stat5fl/fl and Stat5 cKO mice after 0- and 3-h serum starvation was assessed by TUNEL staining. Data are represented as the mean ± SD (n = 3). (A, C, D, F, and G) Bars, 100 µm. Results in all panels are representative of two independent experiments.
Stat5 suppresses MAPK activity by regulating the expression of Dusp1 and Dusp2 in osteoclasts
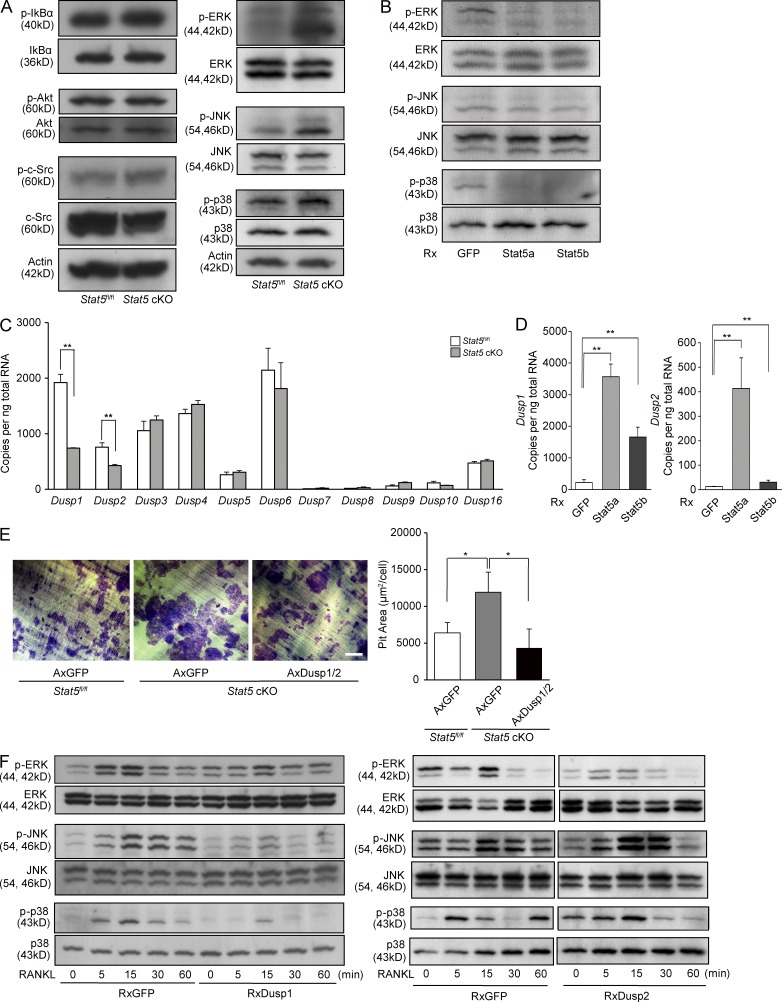
To examine the mechanisms underlying the increase in the bone-resorbing activity of Stat5 cKO osteoclasts, we first analyzed the intracellular signaling pathways. As assessed by Western blotting, the phosphorylation of MAPs, in particular extracellular signal–related kinase 1/2 (ERK1/2), was increased in Stat5 cKO osteoclasts as compared with Stat5fl/fl osteoclasts, whereas the phosphorylation of IκBα, Akt, and c-Src was unchanged by Stat5 deletion (Fig. 4 A). Conversely, retroviral vector–mediated overexpression of Stat5a/b resulted in a diminished MAPK activity (Fig. 4 B).
Figure 4.
Stat5 negatively regulates the activity of MAPKs through the expression of Dusp1 and Dusp2. (A) Whole-cell lysates from Stat5fl/fl and Stat5 cKO osteoclasts were subjected to Western blotting using antibodies against phospho-IκBα, phospho-Akt, phospho–c-Src, phospho-ERK1/2, phospho-JNK, and phopsho-p38. (B) Total cell lysates from WT osteoclasts retrovirally overexpressing Stat5a and Stat5b were analyzed by Western blotting using antibodies against phospho-ERK1/2, phospho-JNK, and phospho-p38. (C) Total RNA was extracted from Stat5fl/fl and Stat5 cKO osteoclasts, and the expression of Dusp1, Dusp2, Dusp3, Dusp4, Dusp5, Dusp6, Dusp7, Dusp8, Dusp9, Dusp10, and Dusp16 was analyzed by quantitative real-time PCR. Data are represented as the mean ± SD (**, P < 0.01; n = 3). (D) Total RNA was prepared from osteoclasts retrovirally overexpressing GFP, Stat5a, or Stat5b, and the expression of Dusp1 and Dusp2 was analyzed by quantitative real-time PCR. Data are represented as the mean ± SD (**, P < 0.01; n = 3). (E) Bone-resorbing activity of Stat5fl/fl (AxGFP) and Stat5 cKO osteoclasts adenovirally transfected with AxGFP and AxDusp1/2 was analyzed by pit formation assay. Representative resorption pits, visualized by toluidine blue staining, are also shown. Graph shows pit resorption area per osteoclast. Data are representative of three independent experiments (*, P < 0.05; n = 3). Bar, 100 µm. (F) Phosphorylation of ERK1/2, JNK, and p38 after RANKL stimulation in WT osteoclasts retrovirally overexpressing Dusp1 or Dusp2 was analyzed by Western blotting. Results are representative of two (A, B, E, and F) or three (C and D) independent experiments.
To uncover the molecular mechanisms of MAPK regulation by Stat5, we undertook comparative microarray screens of Stat5fl/fl and Stat5 cKO osteoclasts. Among the genes whose expression was reduced in Stat5 cKO osteoclasts to less than half of that in Stat5fl/fl osteoclasts, we focused on Dusp2. The Dusps are a heterogeneous group of protein phosphatases that dephosphorylate both phosphotyrosine and phosphoserine/phosphothreonine residues within one substrate. Many of the Dusps serve as MAPK phosphatases (MKPs), which dephosphorylate MAPKs with substrate specificity. Real-time PCR analysis in Stat5fl/fl and Stat5 cKO osteoclasts revealed that among the Dusps with MKP activity, the expression of Dusp1 and Dusp2 was significantly reduced in Stat5 cKO osteoclasts (Fig. 4 C). Overexpression of either Stat5a or Stat5b increased the expression of Dusp1 and Dusp2, with Stat5a exhibiting stronger activity (Fig. 4 D). The increased bone-resorbing activity of Stat5 cKO osteoclasts was disrupted by the adenoviral reintroduction of Dusp1 and Dusp2 (Fig. 4 E). To examine the MKP activity of Dusp1 and Dusp2 in osteoclasts, we overexpressed Dusp1 and Dusp2 using retrovirus vectors. As shown in Fig. 4 F, Dusp1-overexpressing osteoclasts exhibited decreased activity of ERK, JNK, and p38, whereas Dusp2-overexpressing cells exhibited decreased activity of ERK and mildly increased activity of JNK and p38 (Fig. 4 F). These results suggest that the increased activity of MAPKs in Stat5 cKO osteoclasts was caused by the decreased expression of Dusp1 and Dusp2, which in turn resulted in their increased bone-resorbing activity.
IL-3 is a possible stimulator of Stat5 in osteoclasts
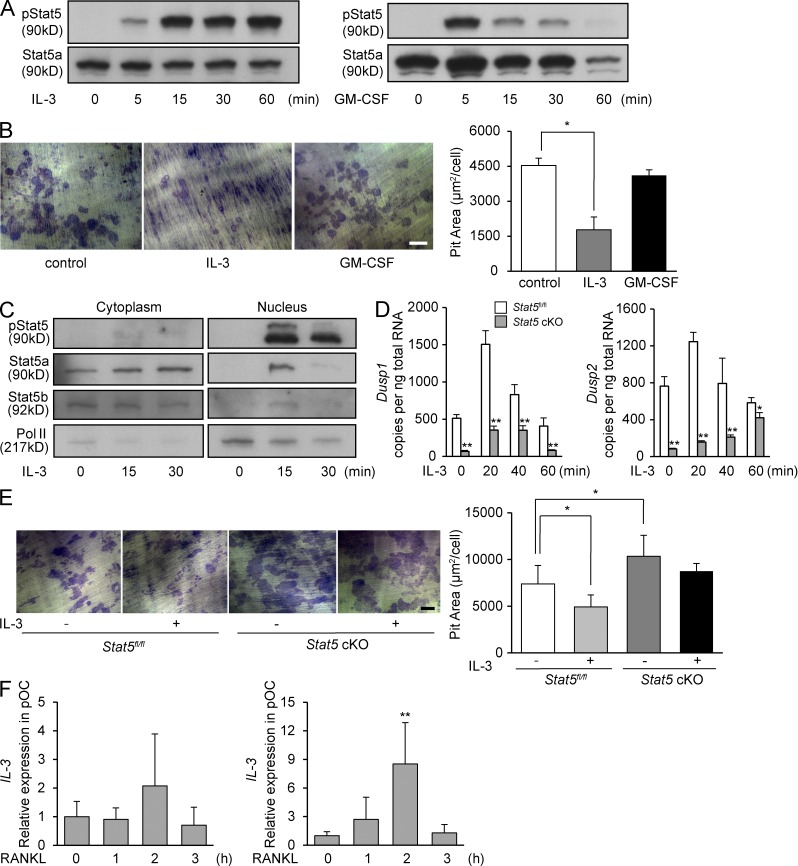
To identify factors that activate Stat5 in osteoclasts, we examined the effects of various cytokines or growth factors on Stat5 phosphorylation. Among the known stimulators of Stat5 (IL-2, IL-3, IL-5, IL-7, GM-CSF, GH, and PRL), IL-3 and GM-CSF induced Stat5 phosphorylation in osteoclasts, although the time course of Stat5 phosphorylation by GM-CSF was more rapid and temporary than that by IL-3 (Fig. 5 A). To determine the effect of these cytokines on bone resorptive activity of osteoclasts, pit formation assay was performed under the presence of IL-3 and GM-CSF. As shown in Fig. 5 B, bone resorption was suppressed by IL-3 but not by GM-CSF stimulation (Fig. 5 B). Therefore, we hypothesized that IL-3–induced activation of Stat5 negatively regulates osteoclast function. Supporting this hypothesis, nuclear translocation of Stat5 was observed in response to IL-3 stimulation (Fig. 5 C). Real-time RT-PCR demonstrated that IL-3 induced a rapid increase in Dusp1 and Dusp2 mRNA in WT osteoclasts, whereas this induction was much reduced in Stat5 cKO osteoclasts (Fig. 5 D). We also found that IL-3 inhibits bone-resorbing activity of Stat5fl/fl osteoclasts, but bone-resorbing activity of Stat5 cKO osteoclasts was not affected by IL-3, as shown in Fig. 5 E. Interestingly, the expression of IL-3 was rapidly induced in osteoclasts in response to RANKL stimulation (Fig. 5 F). These data suggest that the IL-3 produced by osteoclasts activates Stat5 and leads to the suppression of bone resorption by inducing Dusp1 and Dusp2 expression, thus generating a cell-autonomous negative feedback loop in osteoclasts.
Figure 5.
IL-3 may be a stimulator of Stat5 in osteoclasts. (A) WT osteoclasts were stimulated with IL-3 or GM-CSF, and whole-cell lysates were subjected to Western blotting using antibodies against phospho-Stat5 and Stat5a. (B) WT osteoclasts were cultured on dentin slices for 24 h with or without IL-3 and GM-CSF, and the resorption areas were visualized by toluidine blue staining and measured using an image analysis system. Representative resorption pits are shown (left). Graph shows pit resorption area per osteoclast (right). Data are represented as the mean ± SD (*, P < 0.05; n = 3). (C) Osteoclasts from WT mice were stimulated with IL-3 after serum starvation, and cytoplasmic and nuclear fractions were subjected to Western blotting using antibodies against phospho-Stat5, Stat5a, Stat5b, and RNA polymerase II. (D) Osteoclasts from Stat5fl/fl and Stat5 cKO mice were stimulated with IL-3 after serum starvation, and total RNA was extracted. The expression of Dusp1 (left) and Dusp2 (right) was analyzed by quantitative real-time PCR. Data are represented as the mean ± SD (*, P < 0.05; **, P < 0.01; n = 3/group). (E) Stat5fl/fl or Stat5 cKO osteoclasts were cultured on dentin slices for 24 h with or without IL-3, and the resorption areas were visualized by toluidine blue staining and measured using an image analysis system. Representative resorption pits are shown (left). Graph shows pit resorption area per osteoclast (right). Data are represented as the mean ± SD (*, P < 0.05). (B and E) Bars, 100 µm. (F) WT osteoclast precursors and osteoclasts were stimulated with RANKL for the indicated time, total RNA was prepared, and the expression of IL-3 was analyzed by quantitative real-time PCR. Data are represented as the mean ± SD (**, P < 0.01 vs. 0 h; n = 3). Results in all panels are representative of two independent experiments.
DISCUSSION
Multiple previous studies reported important roles for STAT family members in osteoclasts. Takayanagi et al. (2002) reported that β-interferon negatively regulates osteoclast differentiation through the Stat1 pathway, and Abu-Amer (2001) reported that IL-4 abrogates osteoclastogenesis through the Stat6 pathway. The present study demonstrated that Stat5 negatively regulates the bone-resorbing activity of osteoclasts, but it is not essential for their differentiation or survival. Stat family proteins have both redundant and nonredundant functions in many types of cells, and the loss of Stat5 may be compensated by other Stat family members or non-Stat proteins in the regulation of osteoclast differentiation and survival. IL-3, which is induced by RANKL, is a possible mediator of Stat5 activation in osteoclasts.
Stat5 cKO male mice exhibited significantly reduced bone mass at 8 wk of age, but female mice did not show significant difference. The cause of this sex difference is unclear, but Teglund et al. (1998) reported normal growth of Stat5b-deficient female mice. One possible explanation is that the Stat5 signaling pathway is somewhat different between males and females, although further investigation is needed. In an attempt to elucidate the mechanisms by which Stat5 suppresses the bone-resorbing activity of osteoclasts, we demonstrated that the activity of the MAPKs, in particular ERK, was increased in Stat5 KO osteoclasts. Many previous studies have reported the role of ERK in osteoclasts. A recent study using Erk1−/− and Erk2flox/flox mice reported that Erk1 positively regulated osteoclast function (He et al., 2011), providing grounds for our proposal that the increased bone-resorbing activity of Stat5 KO osteoclasts is, at least in part, caused by the increased activity of ERK. Although multiple studies have reported an antiapoptotic effect of ERK in osteoclasts (Miyazaki et al., 2000; Nakamura et al., 2003), we did not find an increased survival of Stat5 KO osteoclasts. This discrepancy may be explained by the duration of ERK phosphorylation and its subcellular localization. Chen et al. (2005) demonstrated that both the duration of ERK activity and localization of ERK are important for the proper function of ERK. They concluded that the kinetics of ERK phosphorylation and the length of time that phospho-ERK is retained in the nucleus are responsible for the pro- versus antiapoptotic effect of ERK. Further studies are needed to precisely determine kinetic activity and localization of ERK in Stat5 KO osteoclasts.
We speculated that the increased MAPK activity observed in Stat5 KO osteoclasts was caused by the absence of the proteins that are induced by Stat5 and affect the phosphorylation status of the MAPKs. As we anticipated, the expression of the MKPs Dusp1 and Dusp2 was reduced in Stat5 cKO osteoclasts. Dusps are a heterogeneous group of protein phosphatases that dephosphorylate both phosphotyrosine and phosphoserine/phosphothreonine residues (Patterson et al., 2009). Some of the Dusp family members are known to serve as MKPs, which, as mentioned above, dephosphorylate MAPKs with substrate specificity. Carlson et al. (2009) reported that Dusp1 negatively regulates the bone-resorbing activity of osteoclasts, whereas the role of Dusp2 in osteoclasts at present remains elusive. Dusp1 is reported to dephosphorylate all of the MAPKs, whereas Dusp2 is induced mainly by ERK. Dusp2 is involved in the anchoring of ERK in the nucleus and suppresses ERK activation (Caunt et al., 2008). In accordance with these studies, the overexpression of Dusp2 in osteoclasts specifically reduced the activity of ERK among various MAPKs. Therefore, the increased MAPK activity in Stat5 KO osteoclasts may be caused by the combined reduction of both Dusp1 and Dusp2.
Among the various factors known to activate Stat5, IL-3 and GM-CSF induced the activation of Stat5 in osteoclasts. However, the duration of Stat5 activation by GM-CSF was much shorter than that by IL-3. Moreover, although IL-3 suppressed the osteoclast bone-resorbing activity, GM-CSF did not affect the bone resorption. Therefore, we speculated that IL-3 exerts a negative effect on bone resorption by Stat5 activation. IL-3 has been reported to suppress osteoclast differentiation by inhibiting the phosphorylation and degradation of IκB (Khapli et al., 2003) or by down-regulating c-Fms (Gupta et al., 2010). IL-3 is produced mainly by T cells after cell activation by antigens and mitogens, suggesting that the IL-3 secreted by activated T cells negatively regulates osteoclast function through the Stat5–Dusp axis. Interestingly, IL-3 expression was increased in osteoclasts in response to RANKL. Therefore, it is also possible that a cell-autonomous IL-3–Stat5–Dusp axis acts as a negative feedback loop in osteoclastic bone resorption. And therefore, IL-3 may have therapeutic potential in bone diseases such as osteoporosis and rheumatoid arthritis, in which osteoclast activity is increased.
A limitation of this study is that the IL-3 effect was assessed only in in vitro cultures of osteoclasts. Further studies are needed to determine whether IL-3 is also implicated in the activation of Stat5 in osteoclasts in vivo. In addition, we did not elucidate which genes regulated by Stat5 are directly responsible for increased activation of Stat5 cKO osteoclasts. Future studies are required to fully understand the mechanisms of transcriptional regulation of osteoclast function. In conclusion, we demonstrated that Stat5 negatively regulates the bone-resorbing function of osteoclasts by promoting Dusp1 and Dusp2 expression, and IL-3 appears to be one of the stimulators of stat5 in osteoclasts.
MATERIALS AND METHODS
Mice.
Stat5fl/fl mice, carrying the Stat5a/b gene with two loxP sequences were generated as previously described (Cui et al., 2004). The mice were on a C57BL/6 background. The presence of the floxed Stat5 gene was determined by PCR around the 5′ loxP site using the primers 5′-GAAAGCATGAAAGGGTTGGAG-3′, 5′-AGCAGCAACCAGAGGACTAC-3′, and 5′-AAGTTATCTCGAGTTAGTCAGG-3′, giving a WT band of 450 bp and a floxed gene product of 200 bp. To generate Stat5 cKO mice, we used cathepsin K–Cre mice (provided by S. Kato, Soma Chuo Hospital, Fukushima, Japan), in which the Cre recombinase gene is knocked into the cathepsin K locus and specifically expressed in osteoclasts (Nakamura et al., 2007). Stat5 cKO mice and normal Stat5fl/fl littermates were generated by mating Cathepsin K–Cre+/−Stat5fl/+ male mice with Cathepsin K–Cre−/−Stat5fl/fl female mice. All animals were housed under specific pathogen–free conditions and treated with humane care under the approval of the Animal Care and Use Committee of the University of Tokyo.
Radiological analyses.
Plain radiographs were taken using a soft x-ray apparatus (CMB-2; SOFTEX), and bone mineral density was measured by DXA using a bone mineral analyzer (PIXImus Densitometer; GE Healthcare). CT scanning of the distal femur was performed using Scan Xmate-L090 (Comscantecno) and was reconstructed into a 3D feature image with 3D-BON (RATOC Systems International).
Serum growth hormone, IGF-I, CTx-I, and osteocalcin measurement.
Blood samples were collected retroorbitally under anesthesia, and serum was obtained using a BD Microtainer. Serum growth hormone was measured using a rat/mouse growth hormone ELISA (EMD Millipore). Serum IGF-I was measured using mouse/rat IGF-I Quantikine ELISA kit (R&D Systems). Serum CTx-I was measured using a RatLaps ELISA kit (Nordic Bioscience Diagnostic A/S). Serum osteocalcin was measured using a mouse osteocalcin EIA kit (Biomedical Technologies). Plasma was obtained using a BD Microtainer.
Histological analyses.
Tissues were fixed in 4% paraformaldehyde/PBS, decalcified in 10% EDTA, embedded in paraffin, and cut into sections of 5-µm thickness. Hematoxylin and eosin (H&E) staining was performed according to the standard procedure. Histomorphometric analysis was performed on undecalcified sections from a point 0.15 mm below the growth plate to 0.6 mm of the primary spongiosa of the proximal tibia. For double labeling, mice were injected subcutaneously with 16 mg/kg body weight of calcein on days 4 and 1 before sacrifice. TRAP-positive cells were stained at pH 5.0 in the presence of L (+)–tartaric acid using naphthol AS-MX phosphate (Sigma-Aldrich) in N,N-dimethyl formamide as the substrate.
Generation of osteoclasts and survival/bone resorption assay.
BM cells were obtained from the femur and tibia of 6-wk-old C57BL/6 mice, and BMDMs were cultured in α-MEM (Gibco) containing 10% FBS (Sigma-Aldrich) in the presence of 100 ng/ml M-CSF (R&D Systems) for 2 d. Osteoclasts were generated by stimulating BMDMs with 10 ng/ml M-CSF and 100 ng/ml RANKL (Wako Pure Chemical Industries, Ltd.) for an additional 4–5 d or by the co-culture system established by Takahashi et al. (1988). The survival assay was performed as follows. After osteoclasts were generated, both RANKL and M-CSF were removed from the culture (time 0), and osteoclasts were cultured for the indicated times. The survival rate of the cells was estimated as the percentage of morphologically intact TRAP+ multinucleated cells compared with those at time 0. Actin ring formation was examined using rhodamine-phalloidin staining. In brief, cells were incubated for 30 min with rhodamine-conjugated phalloidin solution (Molecular Probes). The actin rings formed by osteoclasts were detected with a BZ-8100 fluorescence microscope (KEYENCE). The osteoclast bone resorption assays were performed as previously reported (Miyazaki et al., 2000). In brief, the cells were cultured on dentine slices for 24 h, and the resorption areas were visualized by staining with 1% toluidine blue and then measured using an image analysis system (MicroAnalyzer).
TUNEL staining.
Cells undergoing apoptosis were identified by means of the TdT-mediated dUTP-digoxigenin nick-end labeling (TUNEL) method, which specifically labels the 3′-hydroxyl terminal of DNA strand breaks. The TUNEL staining procedure was performed using In Situ Cell Death Detection kit, Fluorescein (Roche), according to the manufacturer’s recommendation. Apoptotic cells were recognized by fluorescence microscopy.
Expression constructs and gene transduction.
For retrovirus construction, the full-length cDNA of the gene was amplified by PCR using KOD-plus (Takara Bio Inc.), subcloned into Zero Blunt TOPO II vectors (Invitrogen), and inserted into pMX-puro vectors (Kitamura, 1998). 2 × 106 BOSC23 packaging cells were transfected with 6 µg of the vectors using FuGENE 6 (Roche). 24 h later, the medium was replaced with fresh α-MEM/10% FBS, and cells were incubated for an additional 24 h. The supernatant was then collected as retroviral stock after centrifugation at 2,400 rpm for 3 min. 5× 106 BMDMs were incubated with 8 ml of retroviral stock for 5 h in the presence of 6 µg/ml polybrene and 30 ng/ml recombinant mouse M-CSF. After 5 h of retroviral infection, the medium was changed to α-MEM/10% FBS and 100 ng/ml M-CSF, and cells were cultured for an additional 24 h. BMDMs were recovered with trypsin, and puromycin-resistant cells were selected by incubation with α-MEM/10% FBS containing 2 µg/ml puromycin for 2 d and were then used for further experiments. The adenovirus GFP, Stat5a, Stat5b, Dusp1, Dusp2 expression vectors were synthesized using an Adeno-X expression system (Takara Bio Inc.). Viral titers were determined by the end point dilution assay, and the viruses were used at 50 MOI. Adenoviral infection of osteoclasts was performed as previously reported (Tanaka et al., 1998). In short, mouse co-cultures on days 5–6, when the osteoclasts began to appear, were incubated with a small amount of α-MEM containing the recombinant adenoviruses for 1 h at 37°C at the indicated MOI. The cells were washed twice with PBS and further incubated with α-MEM/10% FBS at 37°C. Experiments were performed 24 h after the infection was confirmed.
Real-time PCR analysis.
Total RNA was extracted with ISOGEN (Wako Pure Chemical Industries, Ltd.), and a 1-µg aliquot was reverse transcribed using a QuantiTect Reverse Transcription kit (QIAGEN) to produce single-stranded cDNA. PCR was performed on an ABI Prism 7000 Sequence Detection System (Applied Biosystems) using QuantiTect SYBR Green PCR Master Mix (QIAGEN) according to the manufacturer’s instructions. All reactions were performed in triplicate. After data collection, the mRNA copy number of a specific gene was calculated with a standard curve generated with serially diluted plasmids containing PCR amplicon sequences and then normalized to rodent total RNA with mouse β-actin serving as an internal control. Primer sequences were as follows: β-actin forward, 5′-AGATGTGGATCAGCAAGCA-3′; β-actin reverse, 5′-GCGCAAGTTAGGTTTTGTCA-3′; Stat5a forward, 5′-CCGAAACCTCTGGAATCTGA-3′; Stat5a reverse, 5′-ACGAACTCAGGGACCACTTG-3′; Stat5b forward, 5′-GTGAAGCCACAGATCAAGCA-3′; Stat5b reverse, 5′-TCGGTATCAAGGACGGAGTC-3′; Nfatc1 forward, 5′-TCCGAGAATCGAGATCACCT-3′; Nfatc1 reverse, 5′-AGGGGTCTCTGTAGGCTTCC-3′; Ctsk forward, 5′-GGACCCATCTCTGTGTCCAT-3′; Ctsk reverse, 5′-CCGAGCCAAGAGAGCATATC-3′; Acp5 forward, 5′-CGTCTCTGCACAGATTGCAT-3′; Acp5 reverse, 5′-AACTGCTTTTTGAGCCAGGA-3′; Dusp1 forward, 5′-GAGCTGTGCAGCAAACAGTC-3′; Dusp1 reverse, 5′-CTTCCGAGAAGCGTGATAGG-3′; Dusp2 forward, 5′-ACTGTCCGGATCTGTGCTCT-3′; Dusp2 reverse, 5′-CAGCTGGCAGAGACATTGAG-3′; Dusp3 forward, 5′-TTGAAAGGGCCACAGATTTC-3′; Dusp3 reverse, 5′-AGTTTCACCTTGCCCTCCTT-3′; Dusp4 forward, 5′-CTGTACCTCCCAGCACCAAT-3′; Dusp4 reverse, 5′-GACGGGGATGCACTTGTACT-3′; Dusp5 forward, 5′-TGCACCACCCACCTACACTA-3′; Dusp5 reverse, 5′-AGGACCTTGCCTCCTTCTTC-3′; Dusp6 forward, 5′-TTGAATGTCACCCCCAATTT-3′; Dusp6 reverse, 5′-CATCGTTCATGGACAGGTTG-3′; Dusp7 forward, 5′-TGCCAAGGACTCTACCAACC-3′; Dusp7 reverse, 5′-GAGAGGTTCTGGCTCCAGTG-3′; Dusp8 forward, 5′-GTCCATGAGCCTCTCTCAGC-3′; Dusp8 reverse, 5′-TGAAACGGCTCTCACAGATG-3′; Dusp9 forward, 5′-ACCTTGAGCTGTGGCCTAGA-3′; Dusp9 reverse, 5′-GGGGATCTGCTTGTAGTGGA-3′; Dusp10 forward, 5′-GCGGCAGTACTTTGAAGAGG-3′; Dusp10 reverse, 5′-AGGTTCGGGGAAATAATTGG-3′; Dusp16 forward, 5′-CAGCGAGATGTCCTCAACAA-3′; Dusp16 reverse, 5′-TAAGCACACAGCCATTGGAG-3′; IL-3 forward, 5′-CTGCCTACATCTGCGAATGA-3′; IL-3 reverse, and 5′-TTAGGAGAGACGGAGCCAGA-3′.
Western blotting.
Cells were washed with ice-cold PBS, and proteins were extracted at 4°C with M-PER, an inhibitor cocktail. Equivalent amounts of protein were subjected to SDS-PAGE with 7.5–15% Tris-Glycin gradient gel or 15% Tris-Glycin gel and transferred onto PVDF membranes (Bio-Rad Laboratories, Inc.). After blocking with 6% milk/TBS-T, membranes were incubated with primary antibodies to Stat5a, Stat5b, RNA polymerase (Santa Cruz Biotechnology, Inc.), Cre recombinase (Covance), phospho-Stat5, cleaved caspase-3, ERK1/2, phospho-ERK1/2, JNK, phospho-JNK, p38, phospho-p38, phospho-Akt, Akt, phospho-IκB, IκB, phospho–c-Src (Cell Signaling Technology), c-Src (EMD Millipore), and β-actin (Sigma-Aldrich), followed by HRP-conjugated goat anti–mouse IgG and goat anti–rabbit IgG (Promega). Immunoreactive bands were visualized with ECL (GE Healthcare) according to the manufacturer’s instructions. The blots were stripped by incubating for 20 min in stripping buffer (2% SDS, 100 mM 2-mercaptoethanol, and 62.5 mM Tris-HCl, pH 6.7) at 50°C and then were reprobed with other antibodies.
Microarray analysis.
BM cells were isolated from Stat5fl/fl and Stat5 cKO mice. Osteoclasts were generated from these cells by culture with M-CSF plus RANKL. Total RNA was extracted and microarray analysis was undertaken using the Mouse Oligo chip 24K (TORAY). Data were analyzed using a 3D-Gene Scanner 3000 (TORAY) and deposited in Minimum Information about a Microarray Experiment compliant in Gene Expression Omnibus (GEO accession no. GSE51283).
Data analysis.
Data are presented as the means ± SD. Statistical analyses were performed by two-tailed unpaired Student’s t test. The value of P < 0.05 was considered significant.
Acknowledgments
We thank R. Yamaguchi and H. Kawahara (The University of Tokyo, Bunkyo-ku, Tokyo, Japan) for providing expert technical assistance. Shigeaki Kato provided us with cathepsin K–Cre mice.
This work was supported in part by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan. The research of L. Hennighausen was funded through the Intramural Program of the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health.
Pacific Edit reviewed the manuscript before submission.
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- cKO
- conditional KO
- CT
- computed tomography
- DXA
- dual-energy x-ray absorptiometry
- MAPK
- mitogen-activated protein kinase
- MKP
- MAPK phosphatase
- RANKL
- receptor activator of NF-κB ligand
- TRAP
- tartrate-resistant acid phosphatase
References
- Abu-Amer Y. 2001. IL-4 abrogates osteoclastogenesis through STAT6-dependent inhibition of NF-kappaB. J. Clin. Invest. 107:1375–1385 10.1172/JCI10530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akira S. 1999. Functional roles of STAT family proteins: lessons from knockout mice. Stem Cells. 17:138–146 10.1002/stem.170138 [DOI] [PubMed] [Google Scholar]
- Baron R. 1989. Molecular mechanisms of bone resorption by the osteoclast. Anat. Rec. 224:317–324 10.1002/ar.1092240220 [DOI] [PubMed] [Google Scholar]
- Carlson J., Cui W.G., Zhang Q., Xu X.Q., Mercan F., Bennett A.M., Vignery A. 2009. Role of MKP-1 in osteoclasts and bone homeostasis. Am. J. Pathol. 175:1564–1573 10.2353/ajpath.2009.090035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caunt C.J., Rivers C.A., Conway-Campbell B.L., Norman M.R., McArdle C.A. 2008. Epidermal growth factor receptor and protein kinase C signaling to ERK2: spatiotemporal regulation of ERK2 by dual specificity phosphatases. J. Biol. Chem. 283:6241–6252 10.1074/jbc.M706624200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.R., Plotkin L.I., Aguirre J.I., Han L., Jilka R.L., Kousteni S., Bellido T., Manolagas S.C. 2005. Transient versus sustained phosphorylation and nuclear accumulation of ERKs underlie anti-versus pro-apoptotic effects of estrogens. J. Biol. Chem. 280:4632–4638 10.1074/jbc.M411530200 [DOI] [PubMed] [Google Scholar]
- Cui Y., Riedlinger G., Miyoshi K., Tang W., Li C., Deng C.X., Robinson G.W., Hennighausen L. 2004. Inactivation of Stat5 in mouse mammary epithelium during pregnancy reveals distinct functions in cell proliferation, survival, and differentiation. Mol. Cell. Biol. 24:8037–8047 10.1128/MCB.24.18.8037-8047.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta N., Barhanpurkar A.P., Tomar G.B., Srivastava R.K., Kour S., Pote S.T., Mishra G.C., Wani M.R. 2010. IL-3 inhibits human osteoclastogenesis and bone resorption through downregulation of c-Fms and diverts the cells to dendritic cell lineage. J. Immunol. 185:2261–2272 10.4049/jimmunol.1000015 [DOI] [PubMed] [Google Scholar]
- He Y.Z., Staser K., Rhodes S.D., Liu Y.L., Wu X.H., Park S.J., Yuan J., Yang X.L., Li X.H., Jiang L., et al. 2011. Erk1 positively regulates osteoclast differentiation and bone resorptive activity. PLoS ONE. 6:e24780 10.1371/journal.pone.0024780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennighausen L., Robinson G.W. 2008. Interpretation of cytokine signaling through the transcription factors STAT5A and STAT5B. Genes Dev. 22:711–721 10.1101/gad.1643908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoelbl A., Kovacic B., Kerenyi M.A., Simma O., Warsch W., Cui Y.Z., Beug H., Hennighausen L., Moriggl R., Sexl V. 2006. Clarifying the role of Stat5 in lymphoid development and Abelson-induced transformation. Blood. 107:4898–4906 10.1182/blood-2005-09-3596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilaria R.L., Jr, Van Etten R.A. 1996. P210 and P190(BCR/ABL) induce the tyrosine phosphorylation and DNA binding activity of multiple specific STAT family members. J. Biol. Chem. 271:31704–31710 10.1074/jbc.271.49.31704 [DOI] [PubMed] [Google Scholar]
- Karsenty G., Wagner E.F. 2002. Reaching a genetic and molecular understanding of skeletal development. Dev. Cell. 2:389–406 10.1016/S1534-5807(02)00157-0 [DOI] [PubMed] [Google Scholar]
- Khapli S.M., Mangashetti L.S., Yogesha S.D., Wani M.R. 2003. IL-3 acts directly on osteoclast precursors and irreversibly inhibits receptor activator of NF-kappa B ligand-induced osteoclast differentiation by diverting the cells to macrophage lineage. J. Immunol. 171:142–151 [DOI] [PubMed] [Google Scholar]
- Kitamura T. 1998. New experimental approaches in retrovirus-mediated expression screening. Int. J. Hematol. 67:351–359 10.1016/S0925-5710(98)00025-5 [DOI] [PubMed] [Google Scholar]
- Lacey D.L., Timms E., Tan H.L., Kelley M.J., Dunstan C.R., Burgess T., Elliott R., Colombero A., Elliott G., Scully S., et al. 1998. Osteoprotegerin ligand is a cytokine that regulates osteoclast differentiation and activation. Cell. 93:165–176 10.1016/S0092-8674(00)81569-X [DOI] [PubMed] [Google Scholar]
- Miyazaki T., Katagiri H., Kanegae Y., Takayanagi H., Sawada Y., Yamamoto A., Pando M.P., Asano T., Verma I.M., Oda H., et al. 2000. Reciprocal role of ERK and NF-κB pathways in survival and activation of osteoclasts. J. Cell Biol. 148:333–342 10.1083/jcb.148.2.333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura H., Hirata A., Tsuji T., Yamamoto T. 2003. Role of osteoclast extracellular signal-regulated kinase (ERK) in cell survival and maintenance of cell polarity. J. Bone Miner. Res. 18:1198–1205 10.1359/jbmr.2003.18.7.1198 [DOI] [PubMed] [Google Scholar]
- Nakamura T., Imai Y., Matsumoto T., Sato S., Takeuchi K., Igarashi K., Harada Y., Azuma Y., Krust A., Yamamoto Y., et al. 2007. Estrogen prevents bone loss via estrogen receptor alpha and induction of Fas ligand in osteoclasts. Cell. 130:811–823 10.1016/j.cell.2007.07.025 [DOI] [PubMed] [Google Scholar]
- Patterson K.I., Brummer T., O’Brien P.M., Daly R.J. 2009. Dual-specificity phosphatases: critical regulators with diverse cellular targets. Biochem. J. 418:475–489 [DOI] [PubMed] [Google Scholar]
- Roodman G.D. 2004. Mechanisms of bone metastasis. Discov. Med. 4:144–148 [PubMed] [Google Scholar]
- Seeman E. 2003. Reduced bone formation and increased bone resorption: rational targets for the treatment of osteoporosis. Osteoporos. Int. 14:S2–S8 10.1007/s00198-002-1340-9 [DOI] [PubMed] [Google Scholar]
- Sommer B., Felix R., Sprecher C., Leunig M., Ganz R., Hofstetter W. 2005. Wear particles and surface topographies are modulators of osteoclastogenesis in vitro. J. Biomed. Mater. Res. A. 72A:67–76 10.1002/jbm.a.30202 [DOI] [PubMed] [Google Scholar]
- Suda T., Takahashi N., Udagawa N., Jimi E., Gillespie M.T., Martin T.J. 1999. Modulation of osteoclast differentiation and function by the new members of the tumor necrosis factor receptor and ligand families. Endocr. Rev. 20:345–357 10.1210/er.20.3.345 [DOI] [PubMed] [Google Scholar]
- Takahashi N., Akatsu T., Udagawa N., Sasaki T., Yamaguchi A., Moseley J.M., Martin T.J., Suda T. 1988. Osteoblastic cells are involved in osteoclast formation. Endocrinology. 123:2600–2602 10.1210/endo-123-5-2600 [DOI] [PubMed] [Google Scholar]
- Takayanagi H., Iizuka H., Juji T., Nakagawa T., Yamamoto A., Miyazaki T., Koshihara Y., Oda H., Nakamura K., Tanaka S. 2000. Involvement of receptor activator of nuclear factor kappaB ligand/osteoclast differentiation factor in osteoclastogenesis from synoviocytes in rheumatoid arthritis. Arthritis Rheum. 43:259–269 [DOI] [PubMed] [Google Scholar]
- Takayanagi H., Kim S., Matsuo K., Suzuki H., Suzuki T., Sato K., Yokochi T., Oda H., Nakamura K., Ida N., et al. 2002. RANKL maintains bone homeostasis through c-Fos-dependent induction of interferon-beta. Nature. 416:744–749 10.1038/416744a [DOI] [PubMed] [Google Scholar]
- Tanaka S. 2007. Signaling axis in osteoclast biology and therapeutic targeting in the RANKL/RANK/OPG system. Am. J. Nephrol. 27:466–478 10.1159/000106484 [DOI] [PubMed] [Google Scholar]
- Tanaka S., Takahashi T., Takayanagi H., Miyazaki T., Oda H., Nakamura K., Hirai H., Kurokawa T. 1998. Modulation of osteoclast function by adenovirus vector-induced epidermal growth factor receptor. J. Bone Miner. Res. 13:1714–1720 10.1359/jbmr.1998.13.11.1714 [DOI] [PubMed] [Google Scholar]
- Teglund S., McKay C., Schuetz E., van Deursen J.M., Stravopodis D., Wang D., Brown M., Bodner S., Grosveld G., Ihle J.N. 1998. Stat5a and Stat5b proteins have essential and nonessential, or redundant, roles in cytokine responses. Cell. 93:841–850 10.1016/S0092-8674(00)81444-0 [DOI] [PubMed] [Google Scholar]
- Van Etten R.A. 2007. Aberrant cytokine signaling in leukemia. Oncogene. 26:6738–6749 10.1038/sj.onc.1210758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa A., Guerrini M.M., Cassani B., Pangrazio A., Sobacchi C. 2009. Infantile malignant, autosomal recessive osteopetrosis: the rich and the poor. Calcif. Tissue Int. 84:1–12 10.1007/s00223-008-9196-4 [DOI] [PubMed] [Google Scholar]
- Wakao H., Gouilleux F., Groner B. 1994. Mammary gland factor (MGF) is a novel member of the cytokine regulated transcription factor gene family and confers the prolactin response. EMBO J. 13:2182–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z., Cui Y., Watford W.T., Bream J.H., Yamaoka K., Hissong B.D., Li D., Durum S.K., Jiang Q., Bhandoola A., et al. 2006. Stat5a/b are essential for normal lymphoid development and differentiation. Proc. Natl. Acad. Sci. USA. 103:1000–1005 10.1073/pnas.0507350103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda H., Shima N., Nakagawa N., Yamaguchi K., Kinosaki M., Mochizuki S., Tomoyasu A., Yano K., Goto M., Murakami A., et al. 1998. Osteoclast differentiation factor is a ligand for osteoprotegerin/osteoclastogenesis-inhibitory factor and is identical to TRANCE/RANKL. Proc. Natl. Acad. Sci. USA. 95:3597–3602 10.1073/pnas.95.7.3597 [DOI] [PMC free article] [PubMed] [Google Scholar]