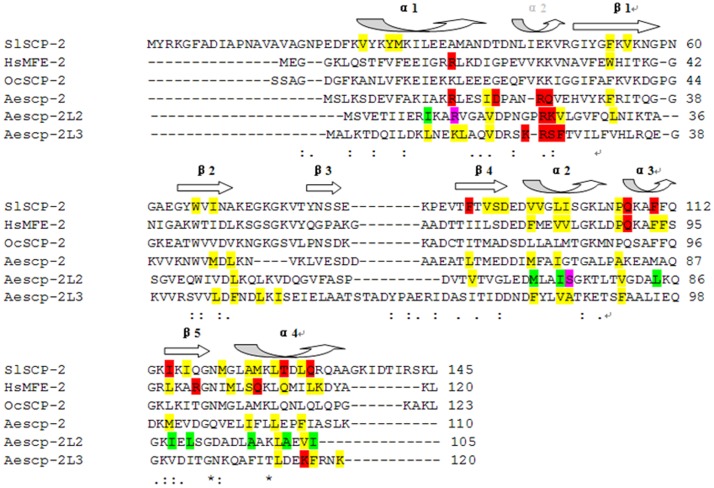
Figure 1. Amino acid sequence alignment of SlSCPx-2 with the SCP-2s, the 3-dimensional structures of which are known.
Red colored residues: coordinating with the polar groups of the ligand. Purple colored residues: coordinating with the polar groups of the second ligand in one chain. Yellow colored residues: interacting directly with the hydrophobic groups of the ligand. Green colored residues: interacting directly with the hydrophobic groups of the second ligand in another chain. Secondary structures are indicated above the amino acid sequences, whereas the secondary structures different from mosquito SCP-2s are in dotted arrow. The alignment was prepared with the program CLUSTALW2 online (http://www.ebi.ac.uk/Tools/msa/clustalw2/).