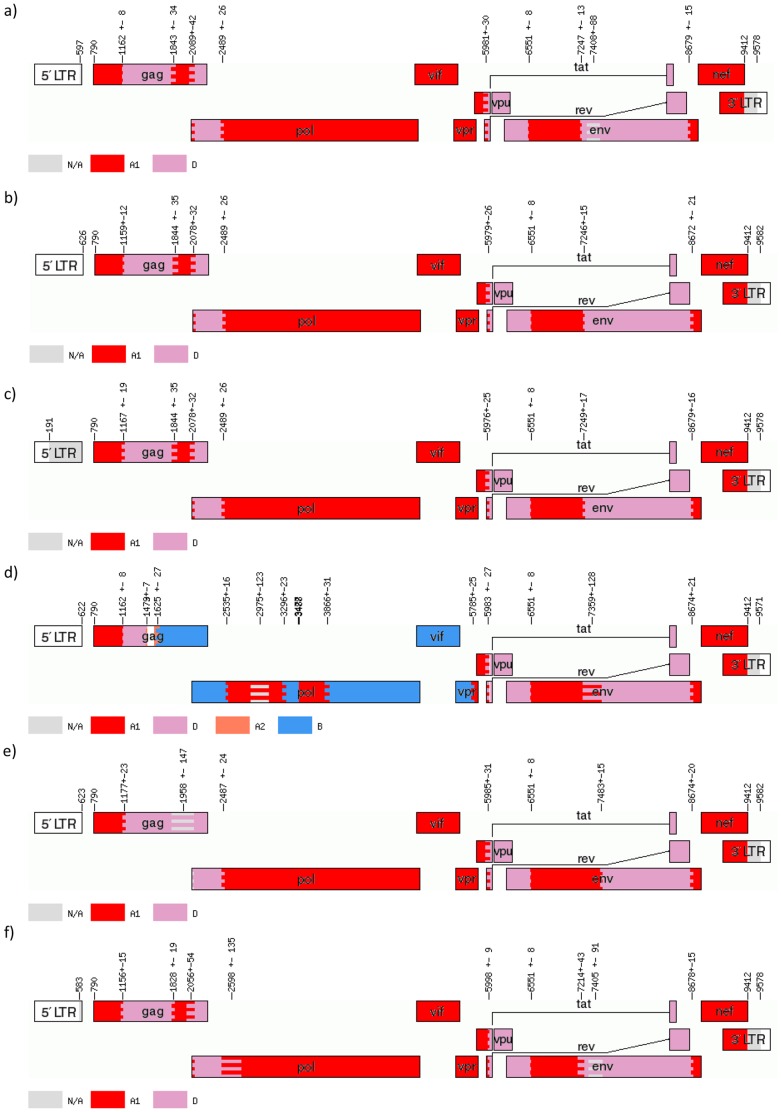
Figure 1. jpHMM analysis of six recombinant HIV-1 sequences.
Putative recombinant HIV-1 sequences were submitted to the online implementation of jpHMM at the GOBICS server. The program used its own stored reference alignment and statistical algorithm to determine subtype classifications, breakpoint locations and 95% confidence intervals. Breakpoint locations and confidence intervals are marked on each plot and are equivalent to HXB2 numbering. In each plot, subtype A1 is represented in red, subtype A2 in coral (plot d only), subtype D in lavender, and subtype B in blue (plot d only). Areas of subtype uncertainty are grey. Five specimens (a, b, c, e, and f) showed largely identical A1/D structures, whereas one specimen (d) showed a complex A1/A2/D/B/U structure. a) Specimen 33365; b) Specimen 8179; c) Specimen 40534; d) Specimen 34567; e) Specimen 11762; f) Specimen 12792.