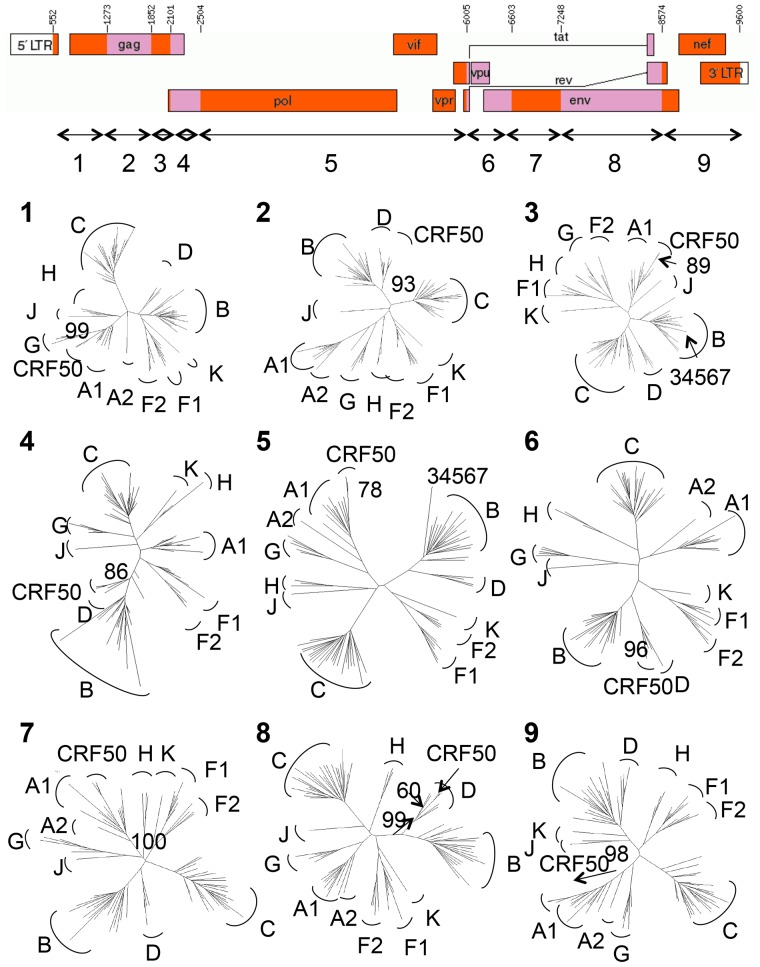
Figure 3. Recombinant map of CRF50_A1D and maximum likelihood phylogenetic trees of non-recombinant fragments.
Maximum likelihood trees of putative non-recombinant fragments from specimens 33365, 8179, 40534 and 34567 drawn using PhyML with PAUP-defined parameters. HIV-1 subtypes used for analysis were A–D, F, G, H, J, K. Numbers indicate bootstrapping support from 1000 replicates (excepting slice 5; 100 replicates). 70% bootstrap support and monophyletic clustering were the criteria for subtype classification. The recombinant map was drawn using the RDT program at Los Alamos. Component subtype fragments are labeled 1–9 on the genome map, corresponding with numbered phylogenetic trees. The genomic regions in which the URF specimen 34567 did not cluster with the CRF50_A1D specimens are indicated in the appropriate trees.