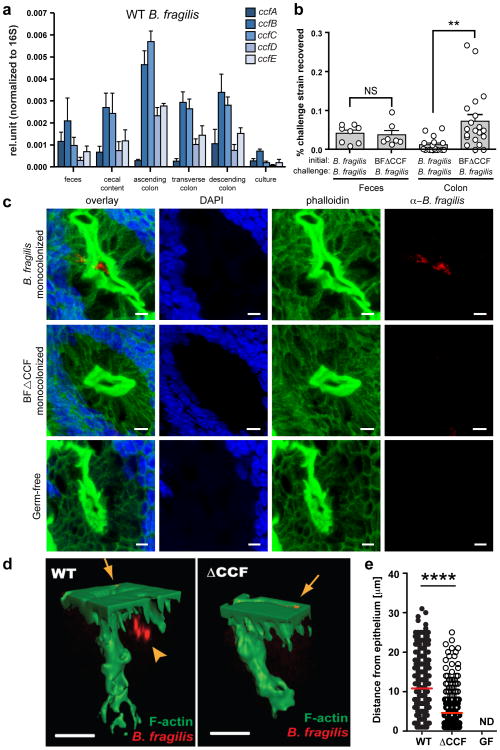
Figure 2. B. fragilis colonization of the colonic crypts is mediated by the CCF system.
a, qRT-PCR of ccf gene expression levels normalized to 16S rRNA (n=3 animals, 2 trials). b, Mice were mono-associated with either WT B. fragilis or B. fragilisΔCCF, and challenged with WT B. fragilis. The percentage of challenge strain was determined in the lumen (feces) and colon after 1 day (n=8 animals/group). c, Confocal micrographs of germ-free, WT B. fragilis or B. fragilisΔCCF mono-associated mice colon whole-mount. Crypts are visualized by DAPI (nuclei, blue) and phalloidin (F-actin, green). Bacteria (red) are stained with IgY polyclonal antibody raised against B. fragilis. Images are representative of 7 different sites analyzed from at least 2 different colons. Scale bar: 5 μm. d, 3D reconstructions of colon crypts from WT B. fragilis or B. fragilisΔCCF mono-associated mice. Bacteria are detected on the apical surface of the epithelium (arrows) and in the crypt space (arrowhead). Scale bar: 10 μm. e, Quantification of bacterial penetration, measured as distance from the epithelial surface per crypt. Error bars indicate standard error of the mean (SEM). NS: not significant. ND: not detected. **p<0.01.****p<0.0001.