Figure 1.
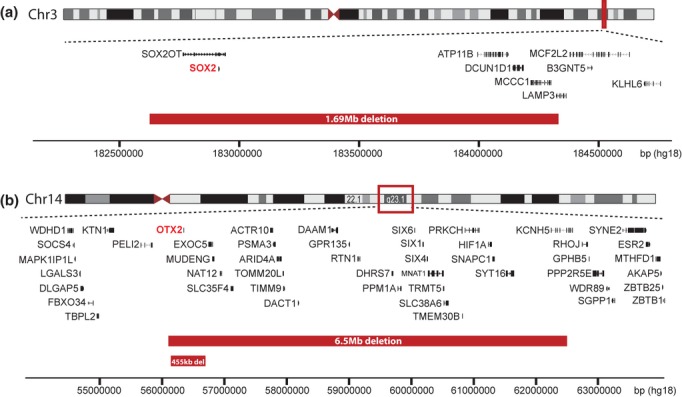
Schematic diagram showing three heterozygous deletions identified by array comparative genomic hybridization (CGH). (A) A de novo 1.69 Mb deletion in CaseID 2850 (chr3:182,649,000–184,339,000 equivalent to hg19 chr3:181,166,306–182,856,306) resulting in a loss of a single copy of SOX2, shown in red. Deletions are marked by red bars. Genomic coordinates are based on the March 2006 Human Genome Assembly (NCBI36/hg18). (B) A 6.5 Mb deletion in CaseID 3000 (chr14:56,094,000–62,594,000; equivalent to hg19 chr14:57,024,247–63,524-247) and a de novo 455 kb deletion in CaseID 3346 (chr14:56,224,000–56,679,000; equivalent to hg19 chr14:57,154,247–57,609,247). Both deletions resulted in a loss of a single copy of OTX2, shown in red.
