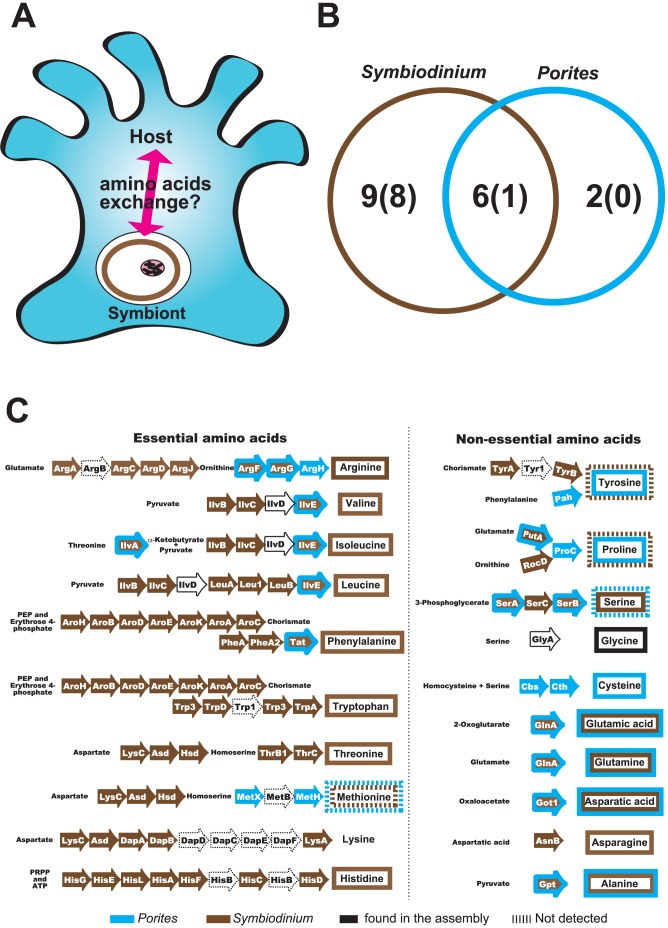
Figure 4. Amino acid metabolism in Porites holobiont.
(A) Schematic drawing of amino acid exchange between a coral polyp and symbiotic Symbiodinium. (B) Summary of amino acid biosynthesic capabilities of Porites (host) and Symbiodinium (symbiont) inferred from KEGG annotation. Numbers indicate how many amino acids can be produced by Porites or Symbiodinium, and numbers in blankets show numbers of essential amino acids. Blue circle indicates Porites and brown circle indicates Symbiodinium, respectively. (C) Detail annotations of amino acid biosynthetic pathways in the Porites holobiont. Arrows indicate enzymes involved in each amino acid biosynthetic pathway. Enzymes that are not annotated as Porites or Symbiodinium are shown with a black line, and those not detected in the transcriptome dataset are shown with dotted lines. Blue indicates enzymes identified in Porites contigs. Brown indicates enzymes detected in Symbiodinium contigs. Brown arrows boxed in blue lines indicate enzymes detected in both Porites and Symbiodinium. Amino acids that can be produced by Symbiodinium are boxed in brown and those that can be produced by Porites are boxed in blue, respectively. Amino acids boxed in dotted lines indicate that biosynthetic capabilities are not clear.