Abstract
Background
Evolution of sturgeons and paddlefishes (order Acipenseriformes) is inherently connected with polyploidization events which resulted in differentiation of ploidy levels and chromosome numbers of present acipenseriform species. Moreover, allopolyploidization as well as autopolyploidization seems to be an ongoing process in these fishes and individuals with abnormal ploidy levels were occasionally observed within sturgeon populations. Here, we reported occurrence of Siberian sturgeon (Acipenser baerii) male with abnormal ploidy level for this species, accessed its ploidy level and chromosome number and investigate its potential sterility or fertility in comparison with normal individuals of sterlet (A. ruthenus), Russian sturgeon (A. gueldenstaedtii) and Siberian sturgeon (A. baerii).
Results
Acipenser ruthenus possessed 120 chromosomes, exhibiting recent diploidy (2n), A. gueldenstaedtii and A. baerii had ~245 chromosomes representing recent tetraploidy (4n), and A. baerii male with abnormal ploidy level had ~ 368 chromosomes, indicating recent hexaploidy (6n). Genealogy assessed from the mtDNA control region did not reveal genome markers of other sturgeon species and this individual was supposed to originate from spontaneous 1.5 fold increment in number of chromosome sets with respect to the number most frequently found in nature for this species. Following hormone stimulation, the spontaneous hexaploid male produced normal sperm with ability for fertilization. Fertilization of A. baerii and A. gueldenstaedtii ova from normal 4n level females with sperm of the hexaploid male produced viable, non-malformed pentaploid (5n) progeny with a ploidy level intermediate to those of the parents.
Conclusion
This study firstly described occurrence of hexaploid individual of A. baerii and confirmed its autopolyploid origin. In addition to that, the first detailed evidence about fertility of spontaneous hexaploid sturgeon was provided. If 1.5 fold increment in number of chromosome sets occurring in diploids, resulted triploids possess odd number of chromosome sets causing their sterility or subfertility due to interference of gametogenesis. In contrast, 1.5 fold increment in number of chromosome sets in naturally tetraploid A. baerii resulted in even number of chromosome sets and therefore in fertility of the hexaploid specimen under study.
Keywords: Acipenseridae, Polyploidy determination, Sperm quality, Autopolyploidization, Triploidization
Background
Evolution of vertebrate genomes was possibly associated with three episodes of whole-genome duplication. The first occurred at the origin of vertebrates and another at the origin of gnathostomes, the 2R hypothesis [1,2]. The third, 3R, is suggested to have occurred in fin-rayed teleostean fishes after their divergence from the earliest lineage of actinopterygians, sturgeon and paddlefish of the extant order Acipenseriformes [3-6].
Sturgeon, paddlefish, the fishes of the genera Psephurus, Polyodon (Acipenseriformes: Polyodontidae), Acipenser, Huso, Scaphirhynchus, and Pseudoscaphirhynchus (Acipenseriformes: Acipenseridae) provide the most remarkable examples of evolution of ploidy levels among vertebrates [7]. Independent of the two ancient genome duplication rounds, several lineage-specific duplication events occurred in sturgeons and paddlefishes [8,9].
Within the most recent vertebrates, sturgeon is among the species with a large number of chromosomes. Nowadays, several well divided groups of acipenseriform species can be recognized depending on DNA content and the number of chromosomes in their cell nuclei. They include species with ~120, ~240 and ~360 chromosomes, corresponding to elevated DNA content [10]. Despite this widely respected division, ploidy status of Acipenseriformes often remains unresolved with conflicting opinions [10,11]. Recent investigations suggest two scales of ploidy levels in Acipenseriformes: the ‘evolutionary scale’, which presumes tetraploid - octaploid - dodecaploid relationships among species, and the ‘recent scale’, which presumes diploid–tetraploid–hexaploid relationships [11].
Probably due to the polyploid nature of their genomes, sturgeon of differing ploidy levels hybridize both in nature [e.g. [12-14] and in captivity [15,16], suggesting mutual fertility of various sturgeon species irrespective of ploidy level. Some authors [17-19] assumed hybrids of parent species having different ploidy levels to be sterile. The same generally applies to fertility of fishes with an odd number of chromosomes, such as autotriploids in which meiosis is seriously affected because three homologous chromosomes cannot pair during the zygotene stage of prophase I, interfering with gonad development and gametogenesis [20].
In the present study, we reported the occurrence of a spontaneous hexaploid male among hatchery stock of Siberian sturgeon, Acipenser baerii, a species of recent tetraploid level 4n with ~245 chromosomes; its experimental hybridization with normal females of A. baerii and A. gueldenstaedtii; and analysis of resulting viable progeny. The main aims of this study were to accessed chromosome number and ploidy level of analyzed individuals and investigate potential sterility or fertility of observed spontaneous hexaploid male of A. baerii.
Results and discussion
Confirmation of ploidy levels of analyzed individuals
Flow cytometry revealed the erythrocyte relative DNA content of the 5 males of A. ruthenus standard to be diploid (2n; Figure 1, peak 1), 1 male and 1 female of A. baerii to be tetraploid (Figure 1, peak 2) and one specimen of A. baerii to be 1.38-fold that of the tetraploids (Figure 1, peak 3). Sperm of analyzed males had average relative DNA content equivalent to haploidy (1.0n; Figure 2, peak 1), diploidy (2.0n; Figure 2, peak 2), and triploidy (3.0n; Figure 2, peak 3), respectively. The erythrocyte relative DNA content of the A. gueldenstaedtii male and female under study was also tetraploid. The coefficient of variation (CV) in both erythrocyte and sperm relative DNA content was below or equal to 2.5% for all specimens. The relative DNA content in juveniles from purebreeding of A. baerii and A gueldenstaedtii (Figure 3a), from the normal x spontaneous hexaploid A. baerii and from the normal A. gueldenstaedtii x spontaneous hexaploid A. baerii hybridizations (Figure 3b) revealed 100% normal ploidy in the purebred juveniles compared to 100% intermediate ploidy level of the F1 hybrid juveniles. This was in agreement with widely accepted theory that hybrids resulted from hybridization between individuals with different chromosome numbers and ploidy levels were considered to have chromosome number and ploidy level intermediate to those of parental individuals [21].
Figure 1.
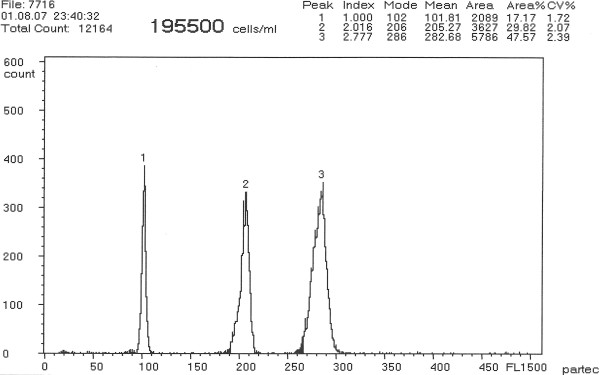
Flow cytometric histogram showing relative DNA content in blood cells of sterlet ( A. ruthenus ; peak 1), Siberian sturgeon ( A. baerii ; peak 2) and spontaneous hexaploid Siberian sturgeon ( A. baerii ; peak 3).
Figure 2.
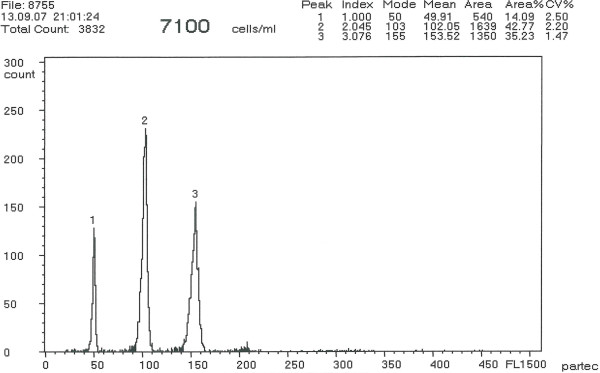
Flow cytometric histogram showing relative DNA content in sperm of sterlet ( A. ruthenus ; peak 1), Siberian sturgeon ( A. baerii ; peak 2) and spontaneous hexaploid Siberian sturgeon ( A. baerii ; peak 3).
Figure 3.
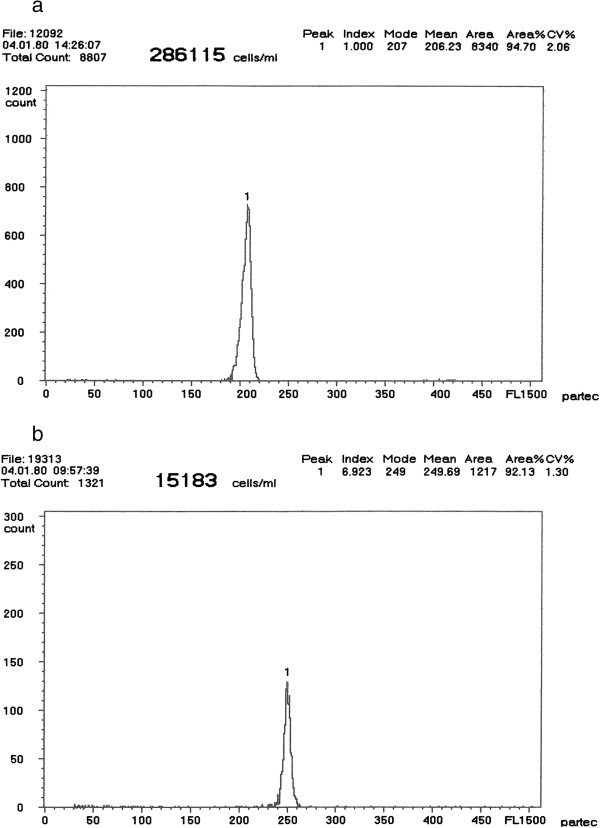
Flow cytometric histogram showing relative DNA content in blood cells of juveniles from A. baerii and A gueldenstaedtii purebreeding (a) and hybridizations of the normal x spontaneous hexaploid A. baerii and the normal A. gueldenstaedtii x spontaneous hexaploid A. baerii (b).
Image cytometry analysis revealed mean erythrocyte nuclear area for A. ruthenus (19.39 ± 1.43a μm2) confirming diploidy, A. baerii (29.95 ± 1.30b μm2) and A. gueldenstaedtii (30.1 ± 1.05b μm2) confirming tetraploidy and one specimen of A. baerii (39.59 ± 3.56c μm2) to be 1.32-fold that of the tetraploids. These results correspond to already published data for sturgeons of higher ploidy levels [22]. These authors confirmed that the erythrocyte nuclear area did not increase linearily with increasing ploidy levels but that the DNA appeared to be more densely packed.
Chromosome analysis of individuals under study revealed A. ruthenus specimens with chromosome number 2n = 120, A. baerii and A. gueldenstaedtii with modal chromosome number 2n ~ 245, while the A. baerii specimen with higher ploidy level exhibited chromosome number 2n ~ 368. Except for microchromosomes, all chromosomes of A. ruthenus could be paired (Figure 4a). Those of A. baerii grouped in quadruplets (Figure 4b), and those of the A. baerii specimen with higher ploidy level grouped into hexaplets (Figure 4d), indicating 1.5 fold increment in number of chromosome sets with respect to the number most frequently found in nature for a given species. If occurring in diploid species, this phenomenon is generally known as triploidization [19]. However, considering the tetraploid status of A. baerii, this phenomenon lead to the formation of hexaploidy.
Figure 4.
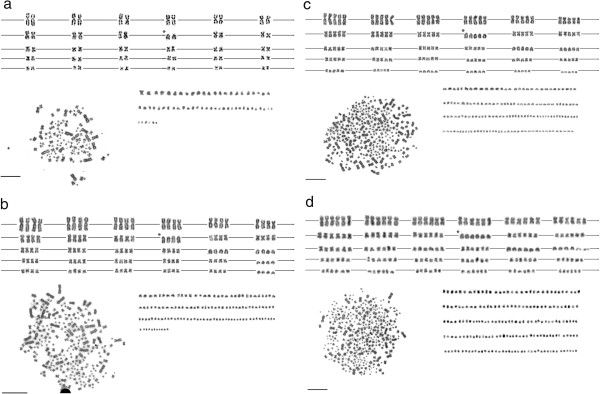
Metaphase cell and corresponding karyotype arranged from Giemsa-stained chromosomes of (a) sterlet Acipenser ruthenus (2n = 120), (b) Siberian sturgeon A. baerii (2n ~ 245), (c) hybrid of A. baerii (2n ~ 245), x spontaneous hexaploid A. baerii (2n ~ 368) resulting in (2n ~ 300) and (d) spontaneous hexaploid A. baerii (2n ~ 368); asterisks denotes „acipenserine“ cytotaxonomic marker, i. e. the group of the largest acrocentric chromosomes. Bars equal 10 μm.
Karyotype analysis of hybrid specimens of the normal x spontaneous hexaploid A. baerii revealed modal number 2n ~ 300 chromosomes, with macrochromosomes that grouped into apparent pentaplets (Figure 4c). The same ploidy level of progeny and karyotype structure resulted from crossing of the normal A. gueldenstadtii female x spontaneous hexaploid A. baerii male.
Ploidy levels of A. baerii and A. gueldenstadtii females used for cross-breeding corresponded to ~245 chromosomes as previously reported for these species [11]. Chromosome number 2n =120 of A. ruthenus used as ploidy standard also corresponded to findings of a number of other cytogenetic studies [23,24]. Ploidy level of progeny resulting from crossing A. baerii and A. gueldenstaedtii with the spontaneous hexaploid A. baerii male was intermediate between the ploidy levels of parents at ~300 chromosomes. This observation was in accordance with findings of Gorshkova et al. [21] and in contrast to the report of Arefjev et al. [25] who demonstrated non-intermediate karyotypes in progeny of hybridization of beluga, Huso huso, and A. gueldenstaedtii. The control mating of normal ploidy level A. baerii and A. gueldenstaedtii yielded progeny of the same ploidy level as the parents. Our results clearly demonstrated that sturgeons with different ploidy levels readily interbreed, as assumed for various combinations of sturgeon species [13].
Confirmation of origin of the hexaploid male A. baerii
The nucleotide sequence of the A. baerii spontaneous hexaploid male was closely similar (99%, E = 0.0) to A. baerii haplotype GenBank: FJ843094.1 [26]. A maximum of three variable sites were detected in common with A. baerii sequences GenBank: EU185048, EU185049 and EU185051 [15]. Similar results were obtained for the A. baerii female. The A. gueldenstaedtii female had mtDNA haplotype closely similar (97%, E = 0.0) to A. gueldenstaedtii haplotype GenBank: FJ843096 [26].
Sturgeons with abnormal ploidy levels have been observed in wild populations [15,27] and in aquaculture [17], and interspecific hybridization has been considered the most probable origin of these. Individuals with abnormal ploidy levels can also arise through autopolyploidization mechanisms such as spontaneous diploidization of the maternal chromosome set (SDM) [28-30] or by polyspermic fertilization [10]. The SDM is a well-known event in fish, and natural autopolyploids resulting from fusion of a haploid sperm pronucleus with an SDM oocyte have been described [28-32].
Because there is no sturgeon species with a higher ploidy level and chromosome number than A. brevirostrum (6n ~372 chromosomes [33]), the hybrid origin of the A. baerii hexaploid might seem improbable. Bearing in mind that SDM of a tetraploid species leads to hexaploidy, the origin of the 1.5 fold increment in number of chromosome sets in genome of the observed specimen might be derived from autopolyploidization, specifically by the junction of pronuclei in a diploid egg, as a result of suppressing the second meiotic division, i.e. by the SDM mechanism. This statement is supported by findings of individuals with abnormal ploidy level among progeny obtained from artificial propagation of A. mikadoi[34]. Furthermore, these individuals showed a modal chromosome number of 360-402 in somatic cells [35], similarly to the hexaploid described in our study. The occurrence of individuals with abnormal ploidy level has also been detected in normal fertilized eggs of the bester (hybrid of H. huso and A. ruthenus) and SDM caused by egg over-ripening was suggested as the most probable cause [36]. Finally, spontaneous autopolyploids of A. transmontanus have also been recently reported [37]. These findings are contradictory to conclusions of Vasiľev [10] who stated that occurrence of spontaneous egg diploidization is close to zero in acipenserids.
Confirmation of fertility of the hexaploid male A. baerii via sperm characteristics
Data on sperm volume per 1 kg MB, numbers of spermatozoa in 1 ml sperm, viability of spermatozoa and their motility and velocity are given in Table 1. The mean velocity of spermatozoa 15 s after activation varied between normal individuals and spontaneous hexaploid male A. baerii (169.66 vs. 152.04 μm s-1, respectively). Velocity of spermatozoa of A. ruthenus was 159 μm s-1, similar to the spontaneous hexaploid A. baerii. The evaluation of spermatozoa motility of hexaploid A. baerii showed normal state of sperm (100 % of motile spermatozoa). Sperm viability assay revealed similar rates of live spermatozoa (98.76 ± 0.36%, 95.93 ± 2.58%, and 93.47 for the A. ruthenus, normal A. baerii, and spontaneous hexaploid A. baerii males, respectively).
Table 1.
Relative volume of sperm kg -1 body mass (M B ), concentration and viability of spermatozoa (spz) and motility and velocity 15 s post activation (mean ± SD) in sperm of A. ruthenus , A. gueldenstaedtii, A. baerii and of the spontaneous hexaploid A. baerii
| Males | No. of Fish | Relative volume of sperm (ml kg -1 M B ) | Concentration (10 9 ml -1 ) | Viability (% live spz) | Motility (% motile spz) | Velocity (μm s -1 ) |
|---|---|---|---|---|---|---|
|
A. ruthenus |
6 |
13.57 ± 4.69 |
1.75 ± 0.46 |
98.76 ± 0.36 |
87.90 ± 12.10 |
159.00 ± 19.25 |
|
A. gueldenstaedtii |
6 |
8.6 ± 4.6 |
1.54 ± 0.80 |
92.56 ± 3.2 |
90.60 ± 9.20 |
165.20 ± 15.72 |
|
A. baerii |
6 |
22.14 ± 7.07 |
1.52 ± 1.88 |
95.93 ± 2.58 |
95.24 ± 9.85 |
169.66 ± 8.69 |
| A. baerii spontaneous hexaploid | 1 | 24.29 | 2.35 | 93.47 | 100.00 | 152.04 |
The observed A. baerii hexaploid produced similar volume of sperm as the diploid specimens. All sperm parameters observed in this experiment were within the range reported by other authors [38-41]. High rates of spermatozoa motility indicated high quality of sperm and corresponded with high viability. The initiation of spermatozoa velocity (in 15 sec) of the hexaploid A. baerii was lower than reported for males of normal ploidy level, as also observed by Pšenička et al. [42]. The authors considered the higher relative DNA content and larger size of heads of the hexaploid A. baerii spermatozoa to be the cause of the difference. Nevertheless this slower movement of spermatozoa did not affect successful fertilization.
Confirmation of fertility of the hexaploid male A. baerii via analyses of its progeny
Experimental fertilization of ova of the normal A. baerii and A. gueldenstaedtii female with sperm of the spontaneous hexaploid A. baerii resulted in hatching rates of 64.16 ± 2.60% and 36.64 ± 2.06%, respectively, similar to that of A. baerii and A. gueldenstaedtii pure-breeding (50.56 ± 8.19% and 39.82 ± 3.64%). No apparent increase in the percent of malformed non-viable larvae was observed in either cross-breeding experiment.
Parental genotypes and observed genotypes of progeny at informative locus Aox45 from experimental crossbreeding (Table 2) confirmed Mendelian segregation patterns in all specimens analyzed. The inheritance of parental alleles of the normal female A. gueldenstaedtii and spontaneous hexaploid male A. baerii were clearly evident at highly informative locus Aox45. The observed genotypes were predominately composed of two maternal and three paternal alleles while locus Afu68 displayed apparently double diploid segregation of alleles. This was in agreement with Ludwig et al. [8], who supposed this locus duplicated in species with ~ 250 chromosomes. The genotypes observed at all analyzed loci are listed in electronic Additional file 1.
Table 2.
Genotyping results at locus Aox45 for hybridization of A. gueldenstaedtii female and A. baerii spontaneous hexaploid male
| Sample | Allele 1 | Allele 2 | Allele 3 | Allele 4 | Allele 5 | Allele 6 | Allele 7 | Allele 8 | Allele 9 |
|---|---|---|---|---|---|---|---|---|---|
|
A. gueldenstaedtii female |
121 |
|
|
|
142 |
145 |
|
|
157 |
|
A. baerii spontaneous hexaploid male |
|
124 |
127 |
136 |
142 |
|
148 |
151 |
|
| progeny 1 |
121 |
124 |
|
|
|
145 |
148 |
151 |
|
| progeny 2 |
121 |
124 |
|
136 |
142 |
|
|
151 |
|
| progeny 3 |
|
|
|
136 |
142 |
|
148 |
|
157 |
| progeny 4 |
|
|
127 |
|
142 |
|
148 |
|
157 |
| progeny 5 |
|
124 |
|
|
142 |
|
|
151 |
157 |
| progeny 6 |
121 |
124 |
127 |
|
|
145 |
|
151 |
|
| progeny 7 |
121 |
124 |
|
|
|
145 |
148 |
151 |
|
| progeny 8 |
|
124 |
|
|
|
145 |
148 |
151 |
157 |
| progeny 9 |
|
|
127 |
|
142 |
|
148 |
|
157 |
| progeny 10 |
|
124 |
127 |
|
142 |
145 |
|
|
157 |
| progeny 11 |
121 |
124 |
127 |
136 |
|
|
|
|
157 |
| progeny 12 |
121 |
124 |
|
|
142 |
|
|
151 |
157 |
| progeny 13 |
|
124 |
127 |
|
142 |
|
|
151 |
157 |
| progeny 14 |
121 |
124 |
|
136 |
142 |
|
|
151 |
|
| progeny 15 |
|
124 |
|
136 |
142 |
|
|
|
157 |
| progeny 16 |
121 |
124 |
|
136 |
142 |
|
148 |
|
|
| progeny 17 |
121 |
|
127 |
|
142 |
145 |
|
151 |
|
| progeny 18 |
|
|
127 |
|
142 |
145 |
148 |
|
157 |
| progeny 19 |
121 |
124 |
|
136 |
142 |
|
|
151 |
|
| progeny 20 |
121 |
124 |
127 |
|
|
145 |
|
151 |
|
| progeny 21 |
|
124 |
127 |
|
142 |
|
|
151 |
157 |
| progeny 22 | 127 | 142 | 148 | 157 |
Fertilization of ova of A. baerii and A. gueldenstaedtii with normal ploidy with sperm of the spontaneous hexaploid A. baerii produced fully viable progeny with ploidy level intermediate to those of the parents. This demonstrated full fertility of the spontaneous hexaploid male. Despite the fact that this specimen has originated by mechanism analogous to spontaneous autotriploidization, where the triploids are generally considered sterile or subfertile due to interference with gametogenesis, meiosis in the hexaploid male A. baerii under study was not affected by this factor. The 1.5 fold increment in number of chromosome sets in tetraploid A. baerii resulted in hexaploidy and homologous chromosomes could apparently group in triplets during the zygotene stage of prophase I during spermatogenesis, as was evident from segregation pattern of microsatellite alleles at representative locus Aox 45 (Table 2).
Conclusion
We confirmed the spontaneous hexaploid status of the examined A. baerii male, i.e. corresponding to ~368 chromosomes of the somatic cells and triploid sperm, a characteristic which has not been reported in other sturgeon species so far.
This study demonstrated that a hexaploid sturgeon male may be fertile owing to its polyploid ancestry and release viable and motile spermatozoa, and that these might have full fertilization potential resulting in a hatching rate similar to that of purebred sturgeon and produce viable progeny with ploidy levels intermediate between those of the parents. Based on these findings, it might be assumed that (auto)polyploids of other sturgeon species could be fertile as well. In light of this phenomenon, it is necessary to evaluate an impact of this event on sturgeon broodstocks in aquaculture, especially if these fishes are used for supportive breeding in reintroduction programs.
Methods
This study was carried out in strict accordance with the Czech Law n. 246/1992 about “Animal welfare”. Authors possess a testimony according to §17 of Law 246/1992 about “Animal welfare”. Protocols have undergone the ethical review process by the University of South Bohemia animal care committee (PP3/FROV/2012; in Czech). Moreover, this was specifically approved by University of South Bohemia animal care committee. All surgery was performed under the clove oil anesthesia, and all efforts were made to minimize suffering.
Fish and breeding details
Parental fish originated from the hatchery of the Faculty of Fisheries and Protection of Waters, University of South Bohemia. One 15-year-old A. baerii male [1.19 m total length (LT), 9.30 kg body mass (MB)], suspected of being spontaneous hexaploid after cytometric examination; one 14-year-old female A. baerii (1.15 m LT, 9.50 kg MB); and one 14-year-old female A. gueldenstaedtii (1.40 m LT, 13.60 kg MB) were used for the experiment. Fifteen-year-old male A. baerii and A. gueldenstaedtii (1.05 m LT and 8.25 kg MB; 0.90m LT and 7.00 kg MB, respectively) were used for controlled breeding with both females. Five sterlet males, Acipenser ruthenus, (0.51 ± 0.15 m LT; 0.65 ± 0.10 kg MB) were used as the diploid standard for flow cytometry and karyotyping. Fish were kept in 5 m3 indoor tanks supplied with re-circulating water at 14°C for 7 days prior to hormone stimulation. Before handling, fish were anesthetized by immersion in 0.07 ml l-1 clove oil [43]. Other 6 males of A. baerii, A. gueldenstaedtii and A. ruthenus were used for comparison of sperm quality parameters with those of hexaploid A. baerii.
Males were stimulated with an intramuscular injection of 4 mg kg-1 MB carp pituitary suspension in physiological saline 36 h before expected sperm sampling [44].
Females were stimulated with an intramuscular injection of 0.5 mg kg-1 MB carp pituitary suspension in physiological saline 42 h before expected ovulation, and again 12 h later with 4.5 mg kg-1 MB of the same suspension [44]. Ovulated eggs for the cross-breeding experiments were collected after microsurgical incision of oviducts as described by Štěch et al.[45]. Fin clips were collected from all parent fish and stored in 96% ethanol.
Sperm quantity and quality
Sperm was collected from the seminal duct, using a 5 mm diameter plastic cannula, into a 100 ml tissue culture flask following the protocol of Gela et al.[44]. Samples were maintained on crushed ice at 0 to 4°C. Sperm volume and spermatozoa concentration, motility, and velocity were assessed according to Linhart et al.[46]. Spermatozoa viability (% live) was determined by epifluorescence microscopy of dual-stained sperm DNA [47]. Wherever applicable, samples were processed in triplicate. The means and standard deviations for sperm characteristics of diploid A. ruthenus, tetraploid A. gueldenstaedtii and tetraploid A. baerii males were calculated from values of each individual.
Fertilization experiment
Eggs of A. baerii and A. gueldenstaedtii were inseminated with sperm from the supposed spontaneous hexaploid A. baerii. Pure-breeding of A. baerii and A. gueldenstaedtii males and females was conducted as control. Eggs were put into plastic beakers in 50 g aliquots, which were placed on a shaking table with constant 200 rpm and 10 mm deflection. Each aliquot was inseminated with 1.5 ml of sperm and activated with 200 ml dechlorinated tap water at 15.0°C. After 2 min, fertilized eggs from each aliquot were separately distributed into 200 cm3 incubator cages, supplied with UV sterilized re-circulating tap water at 15.0°C, 9 mg l-1 O2 in triplicate. During incubation, eggs and hatched larvae were counted, and dead eggs were counted and removed. Hatching rate was computed as described by Linhart et al.[48]. Embryos, swimming-up larvae, and/or early juveniles were sampled for analysis.
Ploidy level analyses
Peripheral blood was collected from the caudal vessel into a heparinized syringe [49], kept at 4°C, and processed immediately with flow cytometry (Partec CCA I; Partec GmbH, Münster, Germany). The ploidy of each adult fish was measured as relative DNA content in blood cells, using 4´,6-diamidino-2-phenylindole (DAPI) first separately for erythrocytes and spermatozoa, and then pooled [48]. Erythrocytes and spermatozoa of a functionally diploid A. ruthenus gave relative DNA content of 2n as the diploid and 1n as the haploid standard. Peripheral bloods of thirty samples of juveniles from each fertilization trial were processed for flow cytometry [50].
For image cytometry, slides were conventionally stained with Giemsa and inspected microscopically (Olympus BHS microscope NCSPlanApo 60x dry objective coupled to a 3CCD Sony DXC-9100P color camera). At least 100 erythrocyte nuclei per specimen were recorded and analyzed by Olympus MicroImage v. 4.0 software. Erythrocyte nuclear area (NA, μm2) was assessed following the protocol in Flajšhans [51]. The NA of functionally diploid A. ruthenus provided the diploid standard.
For karyotyping, metaphase chromosomes were prepared from leucocytes of peripheral blood [52] from all parental individuals. Blood was collected in a heparinized syringe (Zentiva) from the caudal vein and the syringe left in an upright position at 4°C overnight. The sedimented leucocytes and erythrocytes were cultured separately in complete medium (T 199 - Sigma, FBS Superior - Baria, Antibiotic Antimycotic Solution - Sigma, Kanamycin monosulfate - Sigma, LPS - Sigma, PHA H15 - Biomedica, Mercaptoethanol - Sigma) for 6 days at 20°C. The cell suspension was prepared routinely by harvesting cells after colchicine treatment, hypotonization, and fixation. A drop of the cell suspension was placed on a microscope slide, dried, and stained with 5% Giemsa stain buffered to pH 7.0. The chromosomes from five purebred juveniles and five hybrid juveniles from each fertilization trail were prepared according to Völker and Kulmann [53].
Metaphase chromosome plates were examined microscopically (Olympus AX 70) and recorded with an Olympus DP30VW digital camera. Karyotypes were arranged using Ikaros MetaSystems (Metasystems, Germany) software.
Molecular analyses of mitochondrial DNA
The genomic DNA was extracted from fin-clips of all parental individuals using the NucleoSpin®tissue kit (MACHEREY-NAGEL). Amplification followed a standard PCR protocol to amplify a 620 bp mtDNA fragment of the control region[54]. The PCR reaction was carried out under the following conditions: 95°C for 120 s, 5 cycles of 95°C for 60 s, 53 °C for 60 s, and 72°C for 60 s; 30 cycles of 95°C for 30 s, 53°C for 45 s, and 72°C for 60 s; and a final extension at 72°C for 12 min. The PCR products were purified using NucleoSpin® Extract II (MACHEREY-NAGEL) and sequenced in both directions by Macrogen (Seoul, Korea). Sequences were aligned using Geneious 5.4 software [55] and BLASTed against the NCBI nucleotide collection using Mega-BLAST. This NCBI database contains previously published sequences of the control region for most sturgeon species (http://www.ncbi.nlm.nih.gov/).
Fragmentation analyses of microsatellites
The genomic DNA from twenty two swimming-up larvae fixed in 96% ethanol from each trial, as well as from fin-clips of all parent fish was extracted using the NucleoSpin®tissue kit (MACHEREY-NAGEL). Microsatellite DNA fingerprinting of seven microsatellite loci, Afu19, Afu34, Afu39, Afu68[56], Spl101, Spl173[57], and Aox45[58], was used. PCR were performed on a volume of 25 μl, containing 1 U Taq DNA polymerase, 10 pmol of each primer, 10–50 ng DNA, 100 μM of each dNTP, 2.5 mm MgCl2, and 2.5 μl 10 x incubation buffer. Amplifications were performed under the following conditions: one cycle at 94°C for 3min and 25 cycles at 94°C followed by locus-specific annealing conditions: 53°C for locus Aox 45; 57°C for loci Afu 19 Afu 34 Afu 39, Afu 68, Spl 101 and Spl 173; followed by 72°C for 30s and a final extension at 72°C for 10min. The PCR products were inspected on agarose gel, then run in the ABI 3110 DNA analyzer. Genotypes were scored using GeneMapper v4.1 (Applied Biosystems, TM).
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
MH carried out the genotyping and analyses of mtDNA, participated in fertilization experiments and ploidy level assessment by flow cytometry and wrote the manuscript. MHu conducted statistical analyses of mt DNA. PR provided and described karyological data and performed data quality check. MR contributed on karyological analyses and prepared Figure 4. DL participated on microsatellite analyses and calibrated microsatellite scoring. AL performed molecular data check and participated in microsatellite data scoring. MRo supervised artificial reproduction, participated in sperm characterization and evaluation of fertilization experiments. DG was responsible for breading of all parental fishes as well as all progeny. MP provided all sperm characteristics. DB carried out image cytometry slides and contributed to the flow cytometry analyses. MF designed the experiments, carried out flow cytomerty analyses, evaluated all data obtained from flow cytometry and image cytometry, participated in fertilization experimants and performed data check. All authors except MHu read and approved the final version of manuscript.
Supplementary Material
Genotyping results observed at all analyzed loci for hybridization of A. gueldenstaedtii female and A. baerii spontaneous hexaploid male and of A. baerii female and A. baerii spontaneous hexaploid male.
Contributor Information
Miloš Havelka, Email: havelm02@frov.jcu.cz.
Martin Hulák, Email: hulak@frov.jcu.cz.
Petr Ráb, Email: rab@iapg.cas.cz.
Marie Rábová, Email: rabovam@iapg.cas.cz.
Dietmar Lieckfeldt, Email: lieckfeldt@izw-berlin.de.
Arne Ludwig, Email: ludwig@izw-berlin.de.
Marek Rodina, Email: rodina@frov.jcu.cz.
David Gela, Email: gela@frov.jcu.cz.
Martin Pšenička, Email: psenicka@frov.jcu.cz.
Dmytro Bytyutskyy, Email: bytyud00@frov.jcu.cz.
Martin Flajšhans, Email: flajsh@frov.jcu.cz.
Acknowledgements
Dr. M. Hulák passed away on August 29, 2012. His colleagues dedicate this paper to his memory.
Authors would like to acknowledge Dr. Leonardo Congiu for his open approach to constructive discussion during revision of this article and also three anonymous reviewers for their helpful comments. The Lucidus Consultancy is gratefully acknowledged for English correction and suggestions. This study was supported in part by projects CENAKVA CZ.1.05/2.1.00/01.0024, CZ.1.07/2.3.00/30.0049 The Development of Postdoc Positions at the USB, LO1205, GAJU 114/2013/Z and GACR 523/08/0824, although they had no participation in the design of the study nor interpretation of the results. The results of the project LO1205 were obtained with a financial support from the MEYS of the CR under the NPU I program.
References
- McLysaght A, Hokamp K, Wolfe KH. Extensive genomic duplication during early chordate evolution. Nat Genet. 2002;31:200–204. doi: 10.1038/ng884. doi:10.1038/ng884. [DOI] [PubMed] [Google Scholar]
- Panopoulou G, Poustka AJ. Timing and mechanism of ancient vertebrate genome duplications – the adventure of a hypothesis. Trends Genet. 2005;21:559–567. doi: 10.1016/j.tig.2005.08.004. doi:10.1016/.tig.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Ventakhesh B. Evolution and diversity of fish genomes. Curr Opin Genet Dev. 2003;13:588–592. doi: 10.1016/j.gde.2003.09.001. [DOI] [PubMed] [Google Scholar]
- Hoegg S, Brinkmann H, Taylor JS, Meyer A. Phylogenetic timing of the fish-specific genome correlates with the diversification of teleost fish. J Mol Evol. 2004;59:190–203. doi: 10.1007/s00239-004-2613-z. doi:10.1007/s00239-004-2613-z. [DOI] [PubMed] [Google Scholar]
- Volff JN. Genome evolution and biodiversity in teleost fish. Heredity. 2005;94:280–294. doi: 10.1038/sj.hdy.6800635. doi:10.1038/sj.hdy.6800635. [DOI] [PubMed] [Google Scholar]
- Froschauer A, Braasch I, Volff JN. Fish genomes; comparative genomics and vertebrate evolution. Curr Genomics. 2006;7:43–57. doi: 10.2174/138920206776389766. DOI: 1389-2029/06550.00+.00. [DOI] [Google Scholar]
- Nelson JS. Fishes of the World. 4. New York, NY, USA: John Wiley and Sons Inc.; 2006. [Google Scholar]
- Ludwig A, Belfiore NM, Pitra C, Svirsky V, Jenneckens I. Genome duplication events and functional reduction of ploidy levels in sturgeon (Acipenser; Huso and Scaphirhynchus) Genetics. 2001;158:1203–1215. doi: 10.1093/genetics/158.3.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana F, Zane L, Pepe A, Congiu L. In: Fish cytogenetic. Pisano E, Ozof-Costaz C, Foresti F, Kapoor BG, editor. New Hampshire, NH, USA: Science Publisher Inc; 2007. Polyploidy in Acipenseriformes: cytogenetic and molecular approaches; pp. 385–403. [Google Scholar]
- Vasiľev VP. In: Biology, Conservation and Sustainable Development of Sturgeons. Carmona R, Domezain A, García-Gallego M, Hernando JA, Rodríguez F, Ruiz-Rejón M, editor. The Netherlands: Springer Science; 2009. Mechanisms of polyploid evolution in fish: Polyploidy in Sturgeons; pp. 97–117. DOI: 10.1007/978-1-4020-8437-9_6. [Google Scholar]
- Fontana F, Tagliavini J, Congiu C. Sturgeon genetics and cytogenetics: recent advancement and perspectives. Genetic. 2001;111:359–373. doi: 10.1023/a:1013711919443. doi:10.1023/A:1013711919443. [DOI] [PubMed] [Google Scholar]
- Birstein VJ, Poletaev AI, Goncharov BF. DNA content in Eurasian sturgeon species determined by flow cytometry. Cytometry. 1993;14:377–383. doi: 10.1002/cyto.990140406. [DOI] [PubMed] [Google Scholar]
- Birstein VJ, Hanner R, Desalle R. Phylogeny of the Acipenseriformes: Cytogenetic and molecular approaches. Env Biol Fishes. 1997;48:127–156. doi: 10.1023/A:1007366100353. [DOI] [Google Scholar]
- Ludwig A, Debus L, Jenneckens I. A molecular approach for trading control of black caviar. Int Rev Hydrobiology. 2002;87:661–674. doi: 10.1002/1522-2632(200211)87:5/6<661::AID-IROH661>3.0.CO;2-S. DOI: 10.1002/1522 2632(200211)87:5/6B661::AID-IROH661C3.0.CO;2-S. [DOI] [Google Scholar]
- Ludwig A, Lippold S, Debus L, Reinartz R. First evidence of hybridization between endangered starlets (Acipenser ruthenus) and exotic Siberian sturgeons (Acipenser baerii) in the Danube River. Biol Invasions. 2009;11:753–760. doi: 10.1007/s10530-008-9289-z. doi:10.1007/s10530-008-9289-z. [DOI] [Google Scholar]
- Nikolyukin NI. Some observations on the histological structure of the gonads of sturgeon hybrids. Trudy VNIRO. 1964;55:145–157. (in Russian) [Google Scholar]
- Flajšhans M, Vajcová V. Odd ploidy levels in sturgeon suggest a backcross of interspecific hexaploid sturgeon hybrids to evolutionary tetraploid and/or octaploid parental species. Folia Zool. 2000;49:133–138. [Google Scholar]
- Arefjev VA. Sturgeon hybrids: natural reality and practical prospects. Aquac Mag. 1997;23:52–58. [Google Scholar]
- Birstein VJ. In: Endangered Animals: A Reference Guide to Conflicting Issues. Reading RP, Miller B, editor. Westport, CT, USA: Greenwood Press; 2000. Sturgeons and Paddlefishes (Acipenseriformes) pp. 269–278. [Google Scholar]
- Piferrer F, Beaumont A, Falguiere JC, Flajšhans M, Haffray P, Colombo L. Polyploid fish and shellfish: Production; biology and applications to aquaculture for performance improvement and genetic containment. Aquaculture. 2009;239:125–156. doi:10.1016/j.aquaculture.2009.04.036. [Google Scholar]
- Gorshkova G, Gorshkov S, Gordin H, Knibb W. Karyological studies in hybrids of Beluga Huso huso (L.) and the Russian sturgeon Acipenser gueldenstaedtii Brandt. The Israeli J of Aquaculture. 1996;48:35–39. [Google Scholar]
- Bytyutskyy D, Srp J, Flajšhans M. Use of Feulgen image analysis densitometry to study the effect of genome size on nuclear size in polyploid sturgeons. J Appl Ichthyol. 2012;28:704–708. doi: 10.1111/j.1439-0426.2012.02021.x. [DOI] [Google Scholar]
- Ráb P, Arefjev VA, Rábová M. C-banded karyotype of the sterlet; Acipenser ruthenus; from the Danube River. Sturg Quart. 1996;4(Suppl 4):10–12. [Google Scholar]
- Fontana F, Lanfredi M, Chicca M, Congiu L, Tagliavini J, Rossi R. Fluorescent in situ hybridization with rDNA probes on chromosomes of Acipenser ruthenus and Acipenser naccarii (Osteichthyes Acipenseriformes) Genome. 1999;42:1008–1012. doi:10.1139/g99-030. [Google Scholar]
- Arefjev VA, Nikolaev AI. Cytological analysis of the reciprocal hybrids between low- and high-chromosome acipenserids; the great sturgeon; Huso huso (L.); and the Russian sturgeon; Acipenser gueldenstaedti Brandt. Cytologia. 1991;56:495–502. doi: 10.1508/cytologia.56.495. [DOI] [Google Scholar]
- Wang W, Dong Y, Tian ZH, Chen XX, Hu HX. Heteroplasmy in mtDNA control region and phylogenetics of five sturgeons. Zool Res. 2009;30(Suppl 5):487–496. [Google Scholar]
- Dudu A, Suciu R, Paraschiv M, Georgescu SE, Costache M, Berrebi P. Nuclear markers of Danube sturgeon hybridization. Int J Mol Sci. 2011;12:6796–6809. doi: 10.3390/ijms12106796. doi:ijms12106796/ijms12106796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherfas NB, Gomelsky B, Ben–Dom N, Hulata G. Evidence for the heritable nature of spontaneous diploidization in common carp Cyprinus carpio L. eggs. Aquac Res. 1995;26:289–292. doi: 10.1111/j.1365-2109.1995.tb00914.x. doi:10.1111/j.1365-2109.1995.tb00914.x. [DOI] [Google Scholar]
- Aegerter S, Jalabert B. Effects of post-ovulatory oocyte ageing and temperature on egg quality and on the occurrence of triploid fry in rainbow trout; Oncorhynchus mykiss. Aquaculture. 2004;231:59–71. doi: 10.1016/j.aquaculture.2003.08.019. [DOI] [Google Scholar]
- Ezaz MT, McAndrew BJ, Penman DJ. Spontaneous diploidization of the maternal chromosome set in Nile tilapia (Oreochromis niloticus L.) eggs. Aquac Res. 2004;35:271–277. doi: 10.1111/j.1365-2109.2004.01010.x. [DOI] [Google Scholar]
- Centofante L, Bertollo LAC, Moreira-Filho O. Comparative cytogenetics among sympatric species of Characidium (Pisces; Characiformes) Diversity analysis with the description of a ZW sex chromosome system and natural triploidy. Caryologia. 2001;54(Suppl 3):253–260. [Google Scholar]
- Borin LA, Martins-Santos IC, Oliveira C. A natural triploid in Trichomycterus davisi (Siluriformes; Trichomycteridae): mitotic and meiotic characterization by chromosome banding and synaptonemal complex analyses. Genetica. 2002;115:253–258. doi: 10.1023/A:1020667526552. [DOI] [PubMed] [Google Scholar]
- Fontana F, Congiu L, Mudrak VA, Quattro JM, Smith TI, Ware K, Doroshov SI. Evidence of hexaploid karyotype in shortnose sturgeon. Genome. 2008;51:113–119. doi: 10.1139/G07-112. doi:10.1139/G07-112. [DOI] [PubMed] [Google Scholar]
- Zhou H, Fujimoto T, Adachi S, Yamaha E, Arai K. Genome size variation estimated by flow cytometry in Acipenser mikadoi, Huso dauricus in relation to other species of Acipenseriformes. J Appl Ichtyol. 2011;27:484–491. doi: 10.1111/j.1439-0426.2010.01648.x. doi:10.1111/j.1439-0426.2010.01648.x. [DOI] [Google Scholar]
- Zhou H, Fujimoto T, Adachi S, Abe S, Yamaha E, Arai K. Molecular cytogenetic study on the ploidy status in Acipenser mikadoi. J Appl Ichtyol. 2013;29:51–55. doi: 10.1111/jai.12109. doi:10.1111/jai.12109. [DOI] [Google Scholar]
- Omoto N, Maebayashi M, Adachi S, Arai K, Yamauchi K. The influence of oocyte maturational stage on hatching and triploidy rates in hybrid (bester) sturgeon Huso huso x Acipenser ruthenus. Aquaculture. 2005;245:287–294. doi: 10.1016/j.aquaculture.2004.11.008. DOI: 10.1016/j.aquaculture.2004.11.008. [DOI] [Google Scholar]
- Drauch Schreier A, Gille D, Mahardja B, May B. Neutral markers confirm the octaploid origin reveal spontaneous autopolyploidy in white sturgeon, Acipenser transmontanus. J Appl Ichthyol. 2011;27(Suppl 2):24–33. DOI: 10.1111/j.1439-0426.2011.01873.x. [Google Scholar]
- Dettlaff TA, Ginzburg AS, Schmalhausen OI. Sturgeon fishes: developmental biology and aquaculture. Springer – Verlang: Berlin, Germany; 1993. [Google Scholar]
- Hochleitner M. Störe. Österreichischer Agrarverlag: Klosterneuburg, Austria; 1996. (in German) [Google Scholar]
- Dzyuba B, Boryshpolets S, Shaliutina A, Rodina M, Yamaner G, Gela D, Dzyuba V, Linhart O. Spermatozoa motility; cryoresistance; and fertilizing ability in sterlet Acipenser ruthenus during sequential stripping. Aquaculture. 2012;356–357:272–278. doi:10.1016/j.aquaculture.2012.05.006. [Google Scholar]
- Piros B, Glogowski J, Kolman R, Rzemieniecki A, Domagala J, Horváth Á, Urbanyi B, Ciereszko A. Biochemical characterization of Siberian sturgeon Acipenser baerii and sterlet, Acipenser ruthenus, milt plasma and spermatozoa. Fish Physiol Biochem. 2002;26:289–295. doi: 10.1023/A:1026280218957. [DOI] [Google Scholar]
- Pšenička M, Kašpar V, Rodina M, Gela D, Hulák M, Flajšhans M. Comparative study on ultrastructure and motility parameters of spermatozoa of tetraploid and hexaploid Siberian sturgeon Acipenser baerii. J Appl Ichtyol. 2011;27:683–686. doi: 10.1111/j.1439-0426.2011.01685.x. doi:10.1111/j.1439-0426.2011.01685.x. [DOI] [Google Scholar]
- Kolářová J, Velíšek J, Nepejchalová L, Svobodová Z, Kouřil J, Hamáčková J, Máchová J, Piačková V, Hajšlová J, Holadová K, Kocourek V, Klimánková E, Modrá H, Dobšíková R, Groch L, Novotný L. Anestethics for fish - Methodology. Vodňany, Czech Republic: University of South Bohemia in České Budějovice; Research Institute for Fisheries and Hydrobiology; 2007. in Czech. [Google Scholar]
- Gela D, Rodina M, Linhart O. Controlled reproduction of sturgeons (Acipenser) - Methodology. Vodňany, Czech Republic: University of South Bohemia in České Budějovice; Research Institute for Fisheries and Hydrobiology; 2008. in Czech. [Google Scholar]
- Štěch L, Linhart O, Shelton WL, Mims SD. Minimally invasive surgical removal of ovulated eggs of paddlefish (Polyodon spathula) Aqua Int. 1999;7:129–133. doi: 10.1023/A:1009253806766. [DOI] [Google Scholar]
- Linhart O, Rodina M, Cosson J. Cryopreservation of sperm in common carp Cyprinus carpio: sperm motility and hatching success of embryos. Cryobiology. 2000;41:241–250. doi: 10.1006/cryo.2000.2284. doi:10.1006/cryo.2000.2284. [DOI] [PubMed] [Google Scholar]
- Flajšhans M, Cosson J, Rodina M, Linhart O. The application of image cytometry to viability assessment in dual fluorescence-stained fish spermatozoa. Int J Cell Biol. 2004;28:955–959. doi: 10.1016/j.cellbi.2004.07.014. doi:10.1016/j.cellbi.2004.07.014. [DOI] [PubMed] [Google Scholar]
- Linhart O, Rodina M, Flajšhans M, Mavrodiev N, Nebesarova J, Gela D, Kocour M. Studies on sperm of diploid and triploid tench (Tinca tinca L.) Aquac Int. 2006;14:9–25. doi: 10.1007/s10499-005-9010-5. [DOI] [Google Scholar]
- Pravda D, Svobodová Z. In: Veterinary Haematology. Doubek J, Bouda J, Doubek M, Fürll M, Knotková Z, Pejřilová S, editor. Brno, Czech Republic: Noviko; 2003. Haematology of fishes; pp. 381–397. in Czech. [Google Scholar]
- Lecommandeur D, Haffray P, Philippe L. Rapid flow cytometry method for ploidy determination in salmonid eggs. Aquacult Fish Managem. 1994;25:345–350. [Google Scholar]
- Flajšhans M. A model approach to distinguish diploid and triploid fish by means of computer-assisted image analysis. Acta Vet Brno. 1997;66:101–110. doi: 10.2754/avb199766020101. doi:avb199766020101/avb199766020101. [DOI] [Google Scholar]
- Fujiwara A, Nishida-Umehara C, Sakamoto T, Okamoto N, Nakayama I, Abe S. Improved fish lymphocyte culture for chromosome preparation. Genetica. 2001;111:77–89. doi: 10.1023/A:1013788626712. [DOI] [PubMed] [Google Scholar]
- Völker M, Kulmann H. Sequential chromosome banding from single acetic acid fixed embryos of Chromaphyosemion killifishes (Cyprinodontiforme; Nothobranchiidae) Cybium. 2006;30(Suppl 2):171–176. [Google Scholar]
- Mugue NS, Barmintseva AE, Rastorguev SM, Mugue VN, Barminstev VA. Polymorphism of the mitochondrial DNA control region in eight sturgeon species and development of a system for DNA-based species identification. Russ J Genet. 2008;44:793–798. doi: 10.1134/S1022795408070065. DOI: 10.1134/S1022795408070065. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Ashton B, Buxton S, Cheung M, Cooper A, Duran C, Field M, Heled J, Kearse M, Markowitz S, Moir R, Stones-Havas S, Sturrock S, Thierer T, Wilson A. Geneious v 5.4. 2011. Available: http://www.geneious.com.
- May B, Krueger CC, Kincaid HL. Genetic variation at microsatellite loci in sturgeon: primer sequence homology in Acipenser and Scaphirhynchus. Can J Fish Aquat Sci. 1997;54:1542–1547. doi: 10.1139/f97-061. doi:10.1139/cjfas-54-7-1542. [DOI] [Google Scholar]
- McQuown EC, Sloze BL, Sheehan RJ, Rodzen J, Tranah GJ, May B. Microsatellite analysis of genetic variation in sturgeon (Acipenseridae): new primer sequences for Scaphirhynchus and Acipenser. Trans Am Fish Soc. 2000;129:1380–1388. doi: 10.1577/1548-8659(2000)129<1380:MAOGVI>2.0.CO;2. DOI 10.1577/1548-8659(2000)129<1380:MAOGVI>2.0.CO;2. [DOI] [Google Scholar]
- King TL, Lubinski BA, Spidle AP. Microsatellite DNA variation in Atlantic sturgeon (Acipenser oxyrinchus oxyrinchus) and cross-species amplification in the Acipenseridae. Conserv Genet. 2001;2:103–119. doi: 10.1023/A:1011895429669. doi:10.1023/A:1011895429669. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genotyping results observed at all analyzed loci for hybridization of A. gueldenstaedtii female and A. baerii spontaneous hexaploid male and of A. baerii female and A. baerii spontaneous hexaploid male.


