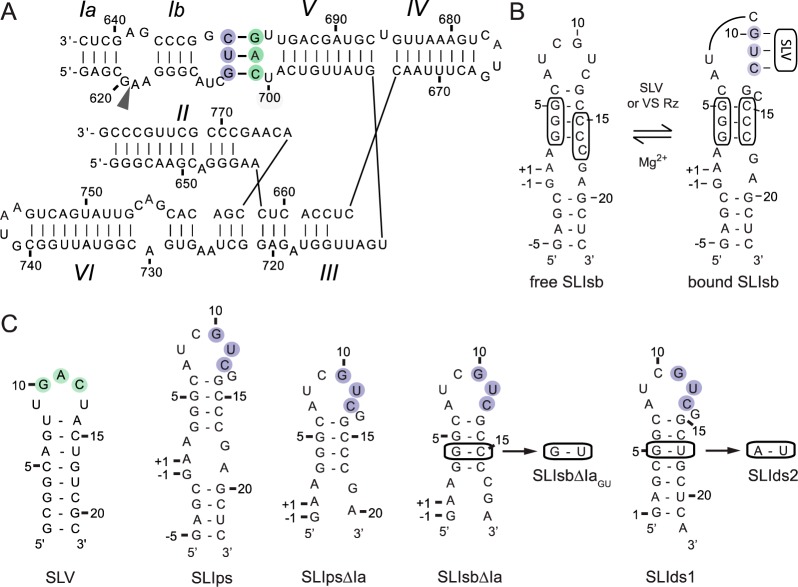
Figure 1.
Primary and secondary structures of RNAs used in this study. (A) The catalytic domain of the Neurospora VS ribozyme containing helical domains II–VI and an SLI substrate (SLIps) containing stems Ia and Ib. The cleavage site is indicated by an arrowhead. The I/V kissing-loop interaction involves Watson–Crick base pairs (thick black lines) between residues G630, U631, and C632 of SLI (shaded in purple) and residues C699, A698, and G697 of SLV (shaded in green). (B) Formation of the I/V kissing-loop is accompanied by a structural rearrangement of the SLI substrate from an unshifted (free SLIsb) to a shifted (bound SLIsb) conformation. (C) The SLV and SLI RNAs used for investigation of SLI/SLV complexes by NMR spectroscopy. In (B) and (C), the cleavage site of the SLI substrates (SLIsb, SLIps, SLIpsΔIa, SLIsbΔIa, and SLIsbΔIaGU) is between residues −1 and +1.